An adaptive threshold in mammalian neocortical evolution
- PMID: 25405475
- PMCID: PMC4236020
- DOI: 10.1371/journal.pbio.1002000
An adaptive threshold in mammalian neocortical evolution
Abstract
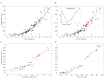
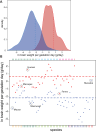
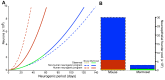
Expansion of the neocortex is a hallmark of human evolution. However, determining which adaptive mechanisms facilitated its expansion remains an open question. Here we show, using the gyrencephaly index (GI) and other physiological and life-history data for 102 mammalian species, that gyrencephaly is an ancestral mammalian trait. We find that variation in GI does not evolve linearly across species, but that mammals constitute two principal groups above and below a GI threshold value of 1.5, approximately equal to 109 neurons, which may be characterized by distinct constellations of physiological and life-history traits. By integrating data on neurogenic period, neuroepithelial founder pool size, cell-cycle length, progenitor-type abundances, and cortical neuron number into discrete mathematical models, we identify symmetric proliferative divisions of basal progenitors in the subventricular zone of the developing neocortex as evolutionarily necessary for generating a 14-fold increase in daily prenatal neuron production, traversal of the GI threshold, and thus establishment of two principal groups. We conclude that, despite considerable neuroanatomical differences, changes in the length of the neurogenic period alone, rather than any novel neurogenic progenitor lineage, are sufficient to explain differences in neuron number and neocortical size between species within the same principal group.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Comment in
-
How folded brains evolved in mammals.PLoS Biol. 2014 Nov 18;12(11):e1002001. doi: 10.1371/journal.pbio.1002001. eCollection 2014 Nov. PLoS Biol. 2014. PMID: 25405604 Free PMC article. No abstract available.
References
-
- Fietz SA, Huttner WB (2011) Cortical progenitor expansion, self-renewal and neurogenesis – a polarized perspective. Curr Opin Neurobiol 21: 23–35. - PubMed
-
- Borrell V, Reillo I (2012) Emerging roles of neural stem cells in cerebral cortex development and evolution. Dev Neurobiol 72: 955–971. - PubMed
-
- Florio M, Huttner WB (2014) Neural progenitors, neurogenesis and the evolution of the neocortex. Development 141: 2182–2194. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

