Genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut
- PMID: 25406499
- PMCID: PMC4246466
- DOI: 10.1186/1471-2164-15-996
Genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut
Erratum in
-
Erratum: genome sequencing of Sporisorium scitamineum provides insights into the pathogenic mechanisms of sugarcane smut.BMC Genomics. 2015 Mar 26;16(1):244. doi: 10.1186/s12864-015-1336-4. BMC Genomics. 2015. PMID: 25880986 Free PMC article. No abstract available.
Abstract
Background: Sugarcane smut can cause losses in cane yield and sugar content that range from 30% to total crop failure. Losses tend to increase with the passage of years. Sporisorium scitamineum is the fungus that causes sugarcane smut. This fungus has the potential to infect all sugarcane species unless a species is resistant to biotrophic fungal pathogens. However, it remains unclear how the fungus breaks through the cell walls of sugarcane and causes the formation of black or gray whip-like structures on the sugarcane plants.
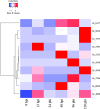
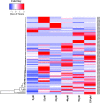
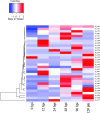
Results: Here, we report the first high-quality genome sequence of S. scitamineum assembled de novo with a contig N50 of 41 kb, a scaffold N50 of 884 kb and genome size 19.8 Mb, containing an estimated 6,636 genes. This phytopathogen can utilize a wide range of carbon and nitrogen sources. A reduced set of genes encoding plant cell wall hydrolytic enzymes leads to its biotrophic lifestyle, in which damage to the host should be minimized. As a bipolar mating fungus, a and b loci are linked and the mating-type locus segregates as a single locus. The S. scitamineum genome has only 6 G protein-coupled receptors (GPCRs) grouped into five classes, which are responsible for transducing extracellular signals into intracellular responses, however, the genome is without any PTH11-like GPCR. There are 192 virulence associated genes in the genome of S. scitamineum, among which 31 expressed in all the stages, which mainly encode for energy metabolism and redox of short-chain compound related enzymes. Sixty-eight candidates for secreted effector proteins (CSEPs) were found in the genome of S. scitamineum, and 32 of them expressed in the different stages of sugarcane infection, which are probably involved in infection and/or triggering defense responses. There are two non-ribosomal peptide synthetase (NRPS) gene clusters that are involved in the generation of ferrichrome and ferrichrome A, while the terpenes gene cluster is composed of three unknown function genes and seven biosynthesis related genes.
Conclusions: As a destructive pathogen to sugar industry, the S. scitamineum genome will facilitate future research on the genomic basis and the pathogenic mechanisms of sugarcane smut.
Figures






References
-
- Mundkur B. Taxonomy of the sugar-cane smuts. Kew Bull. 1939;10(4):525–533.
-
- Hoy J, Hollier C, Fontenot D, Grelen L. Incidence of sugarcane smut in Louisiana and its effect on yield. Plant Dis. 1986;70(1):59–60. doi: 10.1094/PD-70-59. - DOI
-
- Vánky K, Lutz M. Tubisorus, a new genus of smut fungi (Ustilaginomycetes) for Sporisorium pachycarpum. Mycol Balcanica. 2011;8(2):129–135.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

