Use of RAD sequencing for delimiting species
- PMID: 25407078
- PMCID: PMC4815518
- DOI: 10.1038/hdy.2014.105
Use of RAD sequencing for delimiting species
Abstract
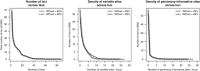
RAD-tag sequencing is a promising method for conducting genome-wide evolutionary studies. However, to date, only a handful of studies empirically tested its applicability above the species level. In this communication, we use RAD tags to contribute to the delimitation of species within a diverse genus of deep-sea octocorals, Chrysogorgia, for which few classical genetic markers have proved informative. Previous studies have hypothesized that single mitochondrial haplotypes can be used to delimit Chrysogorgia species. On the basis of two lanes of Illumina sequencing, we inferred phylogenetic relationships among 12 putative species that were delimited using mitochondrial data, comparing two RAD analysis pipelines (Stacks and PyRAD). The number of homologous RAD loci decreased dramatically with increasing divergence, as >70% of loci are lost when comparing specimens separated by two mutations on the 700-nt long mitochondrial phylogeny. Species delimitation hypotheses based on the mitochondrial mtMutS gene are largely supported, as six out of nine putative species represented by more than one colony were recovered as discrete, well-supported clades. Significant genetic structure (correlating with geography) was detected within one putative species, suggesting that individuals characterized by the same mtMutS haplotype may belong to distinct species. Conversely, three mtMutS haplotypes formed one well-supported clade within which no population structure was detected, also suggesting that intraspecific variation exists at mtMutS in Chrysogorgia. Despite an impressive decrease in the number of homologous loci across clades, RAD data helped us to fine-tune our interpretations of classical mitochondrial markers used in octocoral species delimitation, and discover previously undetected diversity.
Figures


References
-
- Bouchet P, Héros V, Lozouet P, Maestrati P. (2008). A quarter-century of deep-sea malacological exploration in the South and West Pacific: Where do we stand? How far to go? In: Héros V, Cowie RH, Bouchet P (eds). Tropical Deep-Sea Benthos 25, Vol 196, Muséum national d'Histoire naturelle: Paris. pp 9–40.
-
- Calderón I, Garrabou J, Aurelle D. (2006). Evaluation of the utility of COI and ITS markers as tools for population genetic studies of temperate gorgonians. J Exp Mar Biol Ecol 336: 184–197.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

