Sequence-function relationships in folding upon binding
- PMID: 25407143
- PMCID: PMC4315658
- DOI: 10.1002/pro.2605
Sequence-function relationships in folding upon binding
Abstract
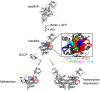
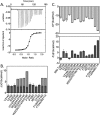
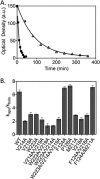
Folding coupled to binding is ubiquitous in biology. Nevertheless, the relationship of sequence to function for protein segments that undergo coupled binding and folding remains to be determined. Specifically, it is not known if the well-established rules that govern protein folding and stability are relevant to ligand-linked folding transitions. Upon small ligand biotinoyl-5'-AMP (bio-5'-AMP) binding the Escherichia coli protein BirA undergoes a disorder-to-order transition that results in formation of a network of packed hydrophobic side chains. Ligand binding is also allosterically coupled to protein association, with bio-5'-AMP binding enhancing the dimerization free energy by -4.0 kcal/mol. Previous studies indicated that single alanine replacements in a three residue hydrophobic cluster that contributes to the larger network disrupt cluster formation, ligand binding, and allosteric activation of protein association. In this work, combined equilibrium and kinetic measurements of BirA variants with alanine substitutions in the entire hydrophobic network reveal large functional perturbations resulting from any single substitution and highly non-additive effects of multiple substitutions. These substitutions also disrupt ligand-linked folding. The combined results suggest that, analogous to protein folding, functional disorder-to-order linked to binding requires optimal packing of the relevant hydrophobic side chains that contribute to the transition. The potential for many combinations of residues to satisfy this requirement implies that, although functionally important, segments of homologous proteins that undergo folding linked to binding can exhibit sequence divergence.
Keywords: folding upon binding; hydrophobic packing; isothermal titration calorimetry; kinetics; sedimentation equilibrium.
© 2014 The Protein Society.
Figures
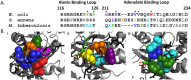
References
-
- Risueno RM, Gil D, Fernandez E, Sanchez-Madrid F, Alarcon B. Ligand-induced conformational change in the T-cell receptor associated with productive immune synapses. Blood. 2005;106:601–608. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

