The immune synapse clears and excludes molecules above a size threshold
- PMID: 25407222
- PMCID: PMC4248232
- DOI: 10.1038/ncomms6479
The immune synapse clears and excludes molecules above a size threshold
Erratum in
-
Corrigendum: The immune synapse clears and excludes molecules above a size threshold.Nat Commun. 2015 Mar 6;6:6109. doi: 10.1038/ncomms7109. Nat Commun. 2015. PMID: 25743540 Free PMC article. No abstract available.
Abstract
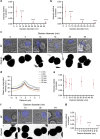
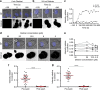
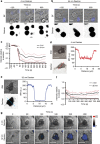
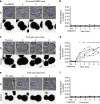
Natural killer cells assess target cell health via interactions at the immune synapse (IS) that facilitates signal integration and directed secretion. Here we test whether the IS also functions as a gasket. Quantitative fluorescence microscopy of nanometer-scale dextrans within synapses formed by various effector-target cell conjugates reveal that molecules are excluded in a size-dependent manner at activating synapses. Dextran sized ≤4 nm move in and out of the IS, but access is significantly reduced (by >50%) for dextran sized 10-13 nm, and dextran ≥32 nm is almost entirely excluded. Depolymerization of F-actin abrogated exclusion. Unexpectedly, larger-sized dextrans are cleared as the IS assembles in a zipper-like manner. Monoclonal antibodies are also excluded from the IS but smaller single-domain antibodies are able to penetrate. Therefore, the IS can clear and exclude molecules above a size threshold, and drugs designed to target synaptic cytokines or cytotoxic proteins must fit these dimensions.
Figures

References
-
- Lanier L. L. NK cell recognition. Annu. Rev. Immunol. 23, 225–274 (2005). - PubMed
-
- Biron C. A., Nguyen K. B., Pien G. C., Cousens L. P. & Salazar-Mather T. P. Natural killer cells in antiviral defense: function and regulation by innate cytokines. Annu. Rev. Immunol. 17, 189–220 (1999). - PubMed
-
- Davis D. M. & Dustin M. L. What is the importance of the immunological synapse? Trends Immunol. 25, 323–327 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

