Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle
- PMID: 25407532
- PMCID: PMC4234836
- DOI: 10.1186/s12711-014-0071-7
Identification of genomic regions associated with inbreeding depression in Holstein and Jersey dairy cattle
Abstract
Background: Inbreeding reduces the fitness of individuals by increasing the frequency of homozygous deleterious recessive alleles. Some insight into the genetic architecture of fitness, and other complex traits, can be gained by using single nucleotide polymorphism (SNP) data to identify regions of the genome which lead to reduction in performance when identical by descent (IBD). Here, we compared the effect of genome-wide and location-specific homozygosity on fertility and milk production traits in dairy cattle.
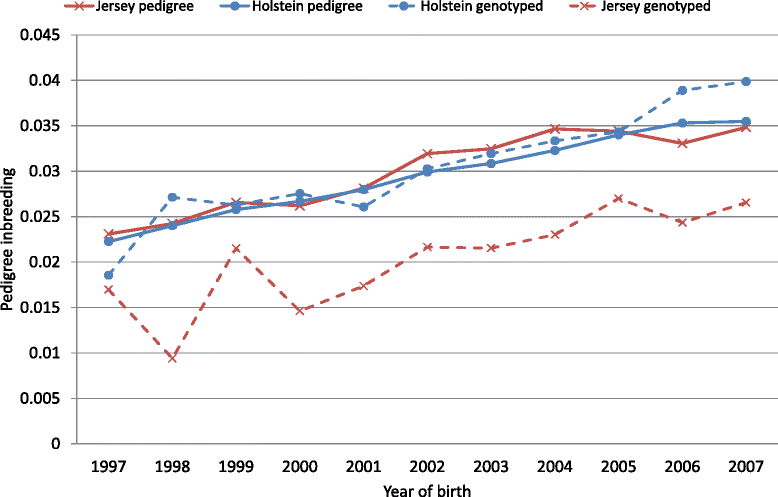
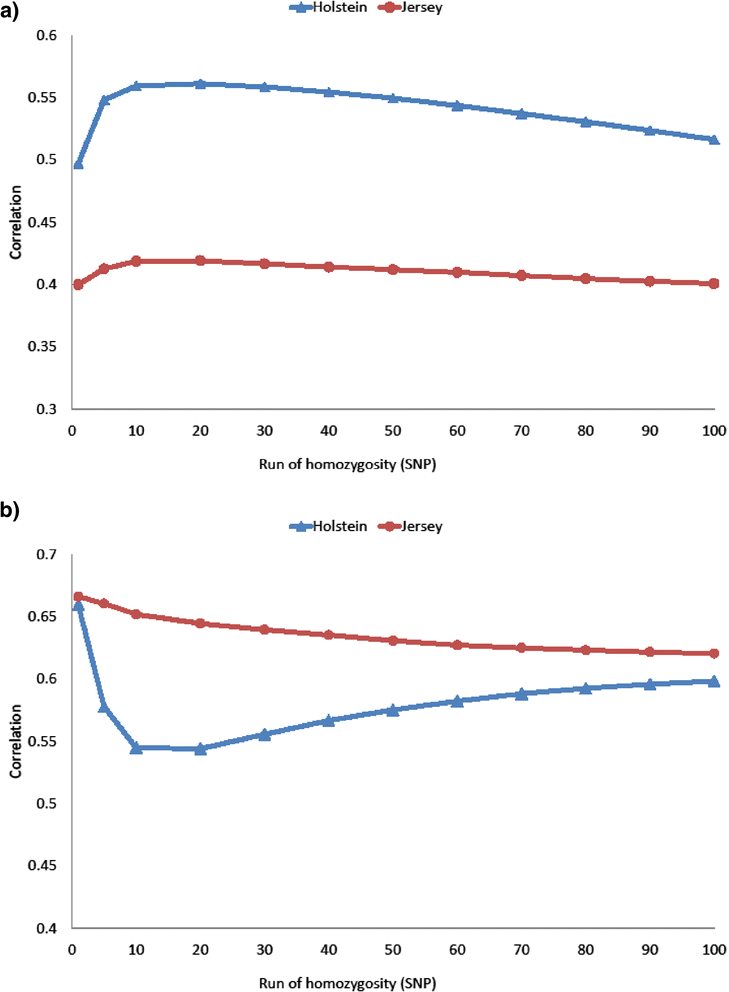
Methods: Genotype data from more than 43 000 SNPs were available for 8853 Holstein and 4138 Jersey dairy cows that were part of a much larger dataset that had pedigree records (338 696 Holstein and 64 049 Jersey animals). Measures of inbreeding were based on: (1) pedigree data; (2) genotypes to determine the realised proportion of the genome that is IBD; (3) the proportion of the total genome that is homozygous and (4) runs of homozygosity (ROH) which are stretches of the genome that are homozygous.
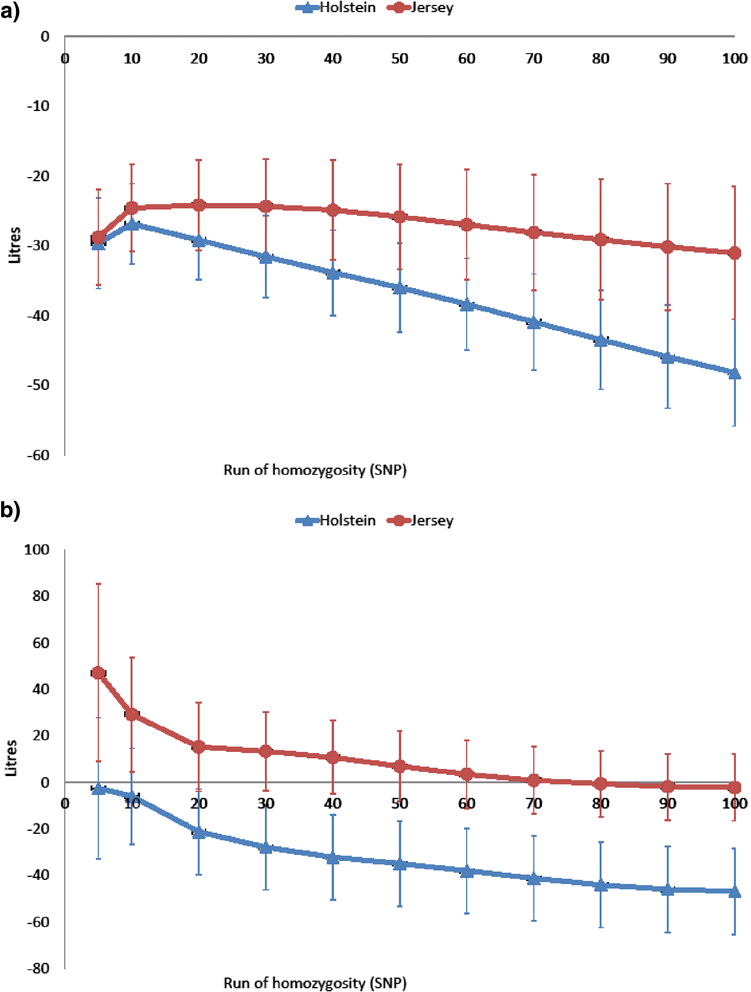
Results: A 1% increase in inbreeding based either on pedigree or genomic data was associated with a decrease in milk, fat and protein yields of around 0.4 to 0.6% of the phenotypic mean, and an increase in calving interval (i.e. a deterioration in fertility) of 0.02 to 0.05% of the phenotypic mean. A genome-wide association study using ROH of more than 50 SNPs revealed genomic regions that resulted in depression of up to 12.5 d and 260 L for calving interval and milk yield, respectively, when completely homozygous.
Conclusions: Genomic measures can be used instead of pedigree-based inbreeding to estimate inbreeding depression. Both the diagonal elements of the genomic relationship matrix and the proportion of homozygous SNPs can be used to measure inbreeding. Longer ROH (>3 Mb) were found to be associated with a reduction in milk yield and captured recent inbreeding independently and in addition to overall homozygosity. Inbreeding depression can be reduced by minimizing overall inbreeding but maybe also by avoiding the production of offspring that are homozygous for deleterious alleles at specific genomic regions that are associated with inbreeding depression.
Figures

References
-
- Keller LF, Waller DM. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17:230–241. doi: 10.1016/S0169-5347(02)02489-8. - DOI
-
- Jarne P, Perdieu M, Pernot A, Delay B, David P. The influence of self-fertilization and grouping on fitness attributes in the freshwater snail Physa acuta: population and individual inbreeding depression. J Evol Biol. 2000;13:645–655. doi: 10.1046/j.1420-9101.2000.00204.x. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

