Generation of multi-gene knockout rabbits using the Cas9/gRNA system
- PMID: 25408890
- PMCID: PMC4230364
- DOI: 10.1186/2045-9769-3-12
Generation of multi-gene knockout rabbits using the Cas9/gRNA system
Abstract
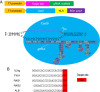
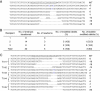
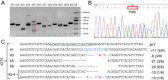
The prokaryotic clustered regularly interspaced short palindromic repeat (CRISPR)-associated system (Cas) is a simple, robust and efficient technique for gene targeting in model organisms such as zebrafish, mice and rats. In this report, we applied CRISPR technology to rabbits by microinjection of Cas9 mRNA and guided RNA (gRNA) into the cytoplasm of pronuclear-stage embryos. We achieved biallelic gene knockout (KO) rabbits by injection of 1 gene (IL2rg) or 2 gene (IL2rg and RAG1) Cas9 mRNA and gRNA with an efficiency of 100%. We also tested the efficiency of multiple gene KOs in early rabbit embryos and found that the efficiency of simultaneous gene mutation on target sites is as high as 100% for 3 genes (IL2rg, RAG1 and RAG2) and 33.3% for 5 genes (IL2rg, RAG1, RAG2, TIKI1 and ALB). Our results demonstrate that the Cas9/gRNA system is a highly efficient and fast tool not only for single-gene editing but also for multi-gene editing in rabbits.
Keywords: Cas9/gRNA system; Multiple-gene knockout; Rabbits.
Figures


References
-
- Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

