The genome of the generalist plant pathogen Fusarium avenaceum is enriched with genes involved in redox, signaling and secondary metabolism
- PMID: 25409087
- PMCID: PMC4237347
- DOI: 10.1371/journal.pone.0112703
The genome of the generalist plant pathogen Fusarium avenaceum is enriched with genes involved in redox, signaling and secondary metabolism
Abstract
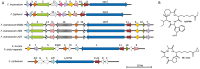
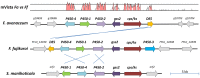
Fusarium avenaceum is a fungus commonly isolated from soil and associated with a wide range of host plants. We present here three genome sequences of F. avenaceum, one isolated from barley in Finland and two from spring and winter wheat in Canada. The sizes of the three genomes range from 41.6-43.1 MB, with 13217-13445 predicted protein-coding genes. Whole-genome analysis showed that the three genomes are highly syntenic, and share>95% gene orthologs. Comparative analysis to other sequenced Fusaria shows that F. avenaceum has a very large potential for producing secondary metabolites, with between 75 and 80 key enzymes belonging to the polyketide, non-ribosomal peptide, terpene, alkaloid and indole-diterpene synthase classes. In addition to known metabolites from F. avenaceum, fuscofusarin and JM-47 were detected for the first time in this species. Many protein families are expanded in F. avenaceum, such as transcription factors, and proteins involved in redox reactions and signal transduction, suggesting evolutionary adaptation to a diverse and cosmopolitan ecology. We found that 20% of all predicted proteins were considered to be secreted, supporting a life in the extracellular space during interaction with plant hosts.
Conflict of interest statement
Figures

Similar articles
-
Fusarium species and enniatin mycotoxins in wheat, durum wheat, triticale and barley harvested in France.Mycotoxin Res. 2019 Nov;35(4):369-380. doi: 10.1007/s12550-019-00363-x. Epub 2019 May 16. Mycotoxin Res. 2019. PMID: 31093880
-
The transcriptome of Fusarium graminearum during the infection of wheat.Mol Plant Microbe Interact. 2011 Sep;24(9):995-1000. doi: 10.1094/MPMI-02-11-0038. Mol Plant Microbe Interact. 2011. PMID: 21585270
-
Fusarium graminearum from expression analysis to functional assays.Methods Mol Biol. 2011;722:79-101. doi: 10.1007/978-1-61779-040-9_6. Methods Mol Biol. 2011. PMID: 21590414
-
Fusarium avenaceum -- the North European situation.Int J Food Microbiol. 2007 Oct 20;119(1-2):17-24. doi: 10.1016/j.ijfoodmicro.2007.07.021. Epub 2007 Jul 31. Int J Food Microbiol. 2007. PMID: 17884217 Review.
-
An update to polyketide synthase and non-ribosomal synthetase genes and nomenclature in Fusarium.Fungal Genet Biol. 2015 Feb;75:20-9. doi: 10.1016/j.fgb.2014.12.004. Epub 2014 Dec 24. Fungal Genet Biol. 2015. PMID: 25543026 Review.
Cited by
-
Chronic Sublethal Aluminum Exposure and Avena fatua Caryopsis Colonization Influence Gene Expression of Fusarium avenaceum F.a.1.Front Microbiol. 2020 Feb 4;11:51. doi: 10.3389/fmicb.2020.00051. eCollection 2020. Front Microbiol. 2020. PMID: 32117103 Free PMC article.
-
Isolation and identification of fungi associated with spoilt fruits vended in Gwagwalada market, Abuja, Nigeria.Vet World. 2017 Apr;10(4):393-397. doi: 10.14202/vetworld.2017.393-397. Epub 2017 Apr 10. Vet World. 2017. PMID: 28507410 Free PMC article.
-
Advances in linking polyketides and non-ribosomal peptides to their biosynthetic gene clusters in Fusarium.Curr Genet. 2019 Dec;65(6):1263-1280. doi: 10.1007/s00294-019-00998-4. Epub 2019 May 28. Curr Genet. 2019. PMID: 31139896 Review.
-
Insights on KP4 Killer Toxin-like Proteins of Fusarium Species in Interspecific Interactions.J Fungi (Basel). 2022 Sep 16;8(9):968. doi: 10.3390/jof8090968. J Fungi (Basel). 2022. PMID: 36135693 Free PMC article.
-
Morphological and Molecular Variation Between Fusarium avenaceum, Fusarium arthrosporioides and Fusarium anguioides Strains.Pathogens. 2018 Nov 29;7(4):94. doi: 10.3390/pathogens7040094. Pathogens. 2018. PMID: 30501049 Free PMC article.
References
-
- Leach MC, Hobbs SLA (2013) Plantwise knowledge bank: delivering plant health information to developing country users. Learned Publ 26: 180–185.
-
- Desjardins AE (2003) Gibberella from a (venaceae) to z (eae). Annu Rev Phytopathol 41: 177–198. - PubMed
-
- Satyaprasad K, Bateman GL, Read PJ (1997) Variation in pathogenicity on potato tubers and sensitivity to thiabendazole of the dry rot fungus Fusarium avenaceum . Potato Res 40: 357–365.
-
- Mercier J, Makhlouf J, Martin RA (1991) Fusarium avenaceum, a pathogen of stored broccoli. Can Plant Dis Surv 71: 161–162.
-
- Sørensen JL, Phipps RK, Nielsen KF, Schroers HJ, Frank J, et al. (2009) Analysis of Fusarium avenaceum metabolites produced during wet apple core rot. J Agricult Food Chem 57: 1632–1639. - PubMed
Publication types
MeSH terms
Associated data
- BioProject/PRJNA253730
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

