Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila
- PMID: 25409104
- PMCID: PMC4402663
- DOI: 10.5665/sleep.4680
Identification of genes associated with resilience/vulnerability to sleep deprivation and starvation in Drosophila
Abstract
Background and study objectives: Flies mutant for the canonical clock protein cycle (cyc(01)) exhibit a sleep rebound that is ∼10 times larger than wild-type flies and die after only 10 h of sleep deprivation. Surprisingly, when starved, cyc(01) mutants can remain awake for 28 h without demonstrating negative outcomes. Thus, we hypothesized that identifying transcripts that are differentially regulated between waking induced by sleep deprivation and waking induced by starvation would identify genes that underlie the deleterious effects of sleep deprivation and/or protect flies from the negative consequences of waking.
Design: We used partial complementary DNA microarrays to identify transcripts that are differentially expressed between cyc(01) mutants that had been sleep deprived or starved for 7 h. We then used genetics to determine whether disrupting genes involved in lipid metabolism would exhibit alterations in their response to sleep deprivation.
Setting: Laboratory.
Patients or participants: Drosophila melanogaster.
Interventions: Sleep deprivation and starvation.
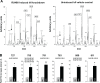
Measurements and results: We identified 84 genes with transcript levels that were differentially modulated by 7 h of sleep deprivation and starvation in cyc(01) mutants and were confirmed in independent samples using quantitative polymerase chain reaction. Several of these genes were predicted to be lipid metabolism genes, including bubblegum, cueball, and CG4500, which based on our data we have renamed heimdall (hll). Using lipidomics we confirmed that knockdown of hll using RNA interference significantly decreased lipid stores. Importantly, genetically modifying bubblegum, cueball, or hll resulted in sleep rebound alterations following sleep deprivation compared to genetic background controls.
Conclusions: We have identified a set of genes that may confer resilience/vulnerability to sleep deprivation and demonstrate that genes involved in lipid metabolism modulate sleep homeostasis.
Keywords: Drosophila melanogaster; lipid metabolism; lipid storage; microarray; sleep homeostasis; transcriptional changes.
© 2015 Associated Professional Sleep Societies, LLC.
Figures




References
-
- Shaw PJ, Cirelli C, Greenspan RJ, Tononi G. Correlates of sleep and waking in Drosophila melanogaster. Science. 2000;287:1834–7. - PubMed
-
- Shaw PJ, Tononi G, Greenspan RJ, Robinson DF. Stress response genes protect against lethal effects of sleep deprivation in Drosophila. Nature. 2002;417:287–91. - PubMed
-
- Everson CA, Bergmann BM, Rechtschaffen A. Sleep deprivation in the rat: III. Total sleep deprivation. Sleep. 1989;12:13–21. - PubMed
-
- Kripke DF, Garfinkel L, Wingard DL, Klauber MR, Marler MR. Mortality associated with sleep duration and insomnia. Archives of general psychiatry. 2002;59:131–6. - PubMed
-
- Van Dongen HP, Maislin G, Mullington JM, Dinges DF. The cumulative cost of additional wakefulness: dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep. 2003;26:117–26. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

