RNA regulons in Hox 5' UTRs confer ribosome specificity to gene regulation
- PMID: 25409156
- PMCID: PMC4353651
- DOI: 10.1038/nature14010
RNA regulons in Hox 5' UTRs confer ribosome specificity to gene regulation
Abstract
Emerging evidence suggests that the ribosome has a regulatory function in directing how the genome is translated in time and space. However, how this regulation is encoded in the messenger RNA sequence remains largely unknown. Here we uncover unique RNA regulons embedded in homeobox (Hox) 5' untranslated regions (UTRs) that confer ribosome-mediated control of gene expression. These structured RNA elements, resembling viral internal ribosome entry sites (IRESs), are found in subsets of Hox mRNAs. They facilitate ribosome recruitment and require the ribosomal protein RPL38 for their activity. Despite numerous layers of Hox gene regulation, these IRES elements are essential for converting Hox transcripts into proteins to pattern the mammalian body plan. This specialized mode of IRES-dependent translation is enabled by an additional regulatory element that we term the translation inhibitory element (TIE), which blocks cap-dependent translation of transcripts. Together, these data uncover a new paradigm for ribosome-mediated control of gene expression and organismal development.
Conflict of interest statement
The authors declare no competing interests.
Figures
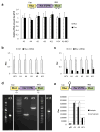




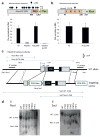
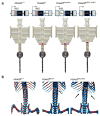
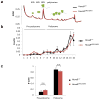
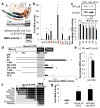




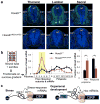
Comment in
-
Molecular biology: Entry signals control development.Nature. 2015 Jan 1;517(7532):24-5. doi: 10.1038/nature14069. Epub 2014 Nov 19. Nature. 2015. PMID: 25409148 Free PMC article.
Similar articles
-
Cis-regulatory RNA elements that regulate specialized ribosome activity.RNA Biol. 2015;12(10):1083-7. doi: 10.1080/15476286.2015.1085149. Epub 2015 Sep 1. RNA Biol. 2015. PMID: 26327194 Free PMC article. Review.
-
Gene- and Species-Specific Hox mRNA Translation by Ribosome Expansion Segments.Mol Cell. 2020 Dec 17;80(6):980-995.e13. doi: 10.1016/j.molcel.2020.10.023. Epub 2020 Nov 16. Mol Cell. 2020. PMID: 33202249 Free PMC article.
-
Molecular biology: Entry signals control development.Nature. 2015 Jan 1;517(7532):24-5. doi: 10.1038/nature14069. Epub 2014 Nov 19. Nature. 2015. PMID: 25409148 Free PMC article.
-
A cross-kingdom internal ribosome entry site reveals a simplified mode of internal ribosome entry.Mol Cell Biol. 2005 Sep;25(17):7879-88. doi: 10.1128/MCB.25.17.7879-7888.2005. Mol Cell Biol. 2005. PMID: 16107731 Free PMC article.
-
Investigation of interactions of polypyrimidine tract-binding protein with artificial internal ribosome entry segments.Biochem Soc Trans. 2005 Dec;33(Pt 6):1483-6. doi: 10.1042/BST0331483. Biochem Soc Trans. 2005. PMID: 16246151 Review.
Cited by
-
Emerging roles of nucleolar and ribosomal proteins in cancer, development, and aging.Cell Mol Life Sci. 2015 Nov;72(21):4015-25. doi: 10.1007/s00018-015-1984-1. Epub 2015 Jul 24. Cell Mol Life Sci. 2015. PMID: 26206377 Free PMC article. Review.
-
The frontier of RNA metamorphosis and ribosome signature in neocortical development.Int J Dev Neurosci. 2016 Dec;55:131-139. doi: 10.1016/j.ijdevneu.2016.02.003. Epub 2016 May 27. Int J Dev Neurosci. 2016. PMID: 27241046 Free PMC article. Review.
-
Ribo-uORF: a comprehensive data resource of upstream open reading frames (uORFs) based on ribosome profiling.Nucleic Acids Res. 2023 Jan 6;51(D1):D248-D261. doi: 10.1093/nar/gkac1094. Nucleic Acids Res. 2023. PMID: 36440758 Free PMC article.
-
Molecular architecture of the ribosome-bound Hepatitis C Virus internal ribosomal entry site RNA.EMBO J. 2015 Dec 14;34(24):3042-58. doi: 10.15252/embj.201592469. Epub 2015 Nov 24. EMBO J. 2015. PMID: 26604301 Free PMC article.
-
Dissection of a rice OsMac1 mRNA 5' UTR to uncover regulatory elements that are responsible for its efficient translation.PLoS One. 2021 Jul 9;16(7):e0253488. doi: 10.1371/journal.pone.0253488. eCollection 2021. PLoS One. 2021. PMID: 34242244 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

