Frequency and fitness consequences of bacteriophage φ6 host range mutations
- PMID: 25409341
- PMCID: PMC4237377
- DOI: 10.1371/journal.pone.0113078
Frequency and fitness consequences of bacteriophage φ6 host range mutations
Abstract
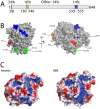
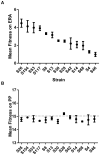
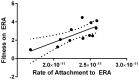
Viruses readily mutate and gain the ability to infect novel hosts, but few data are available regarding the number of possible host range-expanding mutations allowing infection of any given novel host, and the fitness consequences of these mutations on original and novel hosts. To gain insight into the process of host range expansion, we isolated and sequenced 69 independent mutants of the dsRNA bacteriophage Φ6 able to infect the novel host, Pseudomonas pseudoalcaligenes. In total, we found at least 17 unique suites of mutations among these 69 mutants. We assayed fitness for 13 of 17 mutant genotypes on P. pseudoalcaligenes and the standard laboratory host, P. phaseolicola. Mutants exhibited significantly lower fitnesses on P. pseudoalcaligenes compared to P. phaseolicola. Furthermore, 12 of the 13 assayed mutants showed reduced fitness on P. phaseolicola compared to wildtype Φ6, confirming the prevalence of antagonistic pleiotropy during host range expansion. Further experiments revealed that the mechanistic basis of these fitness differences was likely variation in host attachment ability. In addition, using computational protein modeling, we show that host-range expanding mutations occurred in hotspots on the surface of the phage's host attachment protein opposite a putative hydrophobic anchoring domain.
Conflict of interest statement
Figures
References
-
- Christensen KLY, Holman RC, Steiner CA, Sejvar JJ, Stoll BJ, et al. (2009) Infectious disease hospitalizations in the United States. Clinical Infectious Diseases 49: 1025–1035. - PubMed
-
- Armstrong GL, Conn LA, Pinner RW (1999) Trends in infectious disease mortality in the United States during the 20th century. JAMA-Journal of the American Medical Association 281: 61–66. - PubMed
-
- Woolhouse M, Gaunt E (2007) Ecological origins of novel human pathogens. Critical Reviews in Microbiology 33: 231–242. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

