Quantification of histone H3 Lys27 trimethylation (H3K27me3) by high-throughput microscopy enables cellular large-scale screening for small-molecule EZH2 inhibitors
- PMID: 25409661
- PMCID: PMC4361481
- DOI: 10.1177/1087057114559668
Quantification of histone H3 Lys27 trimethylation (H3K27me3) by high-throughput microscopy enables cellular large-scale screening for small-molecule EZH2 inhibitors
Abstract
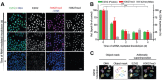
EZH2 inhibition can decrease global histone H3 lysine 27 trimethylation (H3K27me3) and thereby reactivates silenced tumor suppressor genes. Inhibition of EZH2 is regarded as an option for therapeutic cancer intervention. To identify novel small-molecule (SMOL) inhibitors of EZH2 in drug discovery, trustworthy cellular assays amenable for phenotypic high-throughput screening (HTS) are crucial. We describe a reliable approach that quantifies changes in global levels of histone modification marks using high-content analysis (HCA). The approach was validated in different cell lines by using small interfering RNA and SMOL inhibitors. By automation and miniaturization from a 384-well to 1536-well plate, we demonstrated its utility in conducting phenotypic HTS campaigns and assessing structure-activity relationships (SAR). This assay enables screening of SMOL EZH2 inhibitors and can advance the mechanistic understanding of H3K27me3 suppression, which is crucial with regard to epigenetic therapy. We observed that a decrease in global H3K27me3, induced by EZH2 inhibition, comprises two distinct mechanisms: (1) inhibition of de novo DNA methylation and (II) inhibition of dynamic, replication-independent H3K27me3 turnover. This report describes an HCA assay for primary HTS to identify, profile, and optimize cellular active SMOL inhibitors targeting histone methyltransferases, which could benefit epigenetic drug discovery.
Keywords: EZH2; KMT6; chromatin modulators; high-content analysis; histone methyltransferase.
© 2014 Society for Laboratory Automation and Screening.
Conflict of interest statement
Figures



Similar articles
-
Identification of catalytic and non-catalytic activity inhibitors against PRC2-EZH2 complex through multiple high-throughput screening campaigns.Chem Biol Drug Des. 2020 Oct;96(4):1024-1051. doi: 10.1111/cbdd.13702. Epub 2020 May 25. Chem Biol Drug Des. 2020. PMID: 32394628
-
Yin Yang 1-mediated epigenetic silencing of tumour-suppressive microRNAs activates nuclear factor-κB in hepatocellular carcinoma.J Pathol. 2016 Apr;238(5):651-64. doi: 10.1002/path.4688. J Pathol. 2016. PMID: 26800240
-
Enhancer of zeste homolog 2 depletion induces cellular senescence via histone demethylation along the INK4/ARF locus.Int J Biochem Cell Biol. 2015 Aug;65:104-12. doi: 10.1016/j.biocel.2015.05.011. Epub 2015 May 22. Int J Biochem Cell Biol. 2015. PMID: 26004298
-
Targeting histone methyltransferase EZH2 as cancer treatment.J Biochem. 2014 Nov;156(5):249-57. doi: 10.1093/jb/mvu054. Epub 2014 Aug 31. J Biochem. 2014. PMID: 25179367 Review.
-
EZH2 as a potential target in cancer therapy.Epigenomics. 2014 Jun;6(3):341-51. doi: 10.2217/epi.14.23. Epigenomics. 2014. PMID: 25111487 Review.
Cited by
-
Uncovering epigenetic landscape: a new path for biomarkers identification and drug development.Mol Biol Rep. 2020 Nov;47(11):9097-9122. doi: 10.1007/s11033-020-05916-3. Epub 2020 Oct 21. Mol Biol Rep. 2020. PMID: 33089404 Review.
-
A miniaturized mode-of-action profiling platform enables high throughput characterization of the molecular and cellular dynamics of EZH2 inhibition.Sci Rep. 2024 Jan 19;14(1):1739. doi: 10.1038/s41598-023-50964-x. Sci Rep. 2024. PMID: 38242973 Free PMC article.
-
Screening for Small-Molecule Inhibitors of Histone Methyltransferases.Methods Mol Biol. 2022;2529:477-490. doi: 10.1007/978-1-0716-2481-4_20. Methods Mol Biol. 2022. PMID: 35733027
-
Cell-based assays to support the profiling of small molecules with histone methyltransferase and demethylase modulatory activity.Drug Discov Today Technol. 2015 Nov;18:9-17. doi: 10.1016/j.ddtec.2015.10.004. Epub 2015 Nov 3. Drug Discov Today Technol. 2015. PMID: 26723887 Free PMC article. Review.
-
Improving drug discovery using image-based multiparametric analysis of the epigenetic landscape.Elife. 2019 Oct 22;8:e49683. doi: 10.7554/eLife.49683. Elife. 2019. PMID: 31637999 Free PMC article.
References
-
- Shi Y., Lan F., Matson C., et al. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog LSD1. Cell 2004, 119, 941–953. - PubMed
-
- Shi Y., Whetstine J. R. Dynamic Regulation of Histone Lysine Methylation by Demethylases. Mol. Cell. 2007, 25, 1–14. - PubMed
-
- Agger K., Cloos P. A., Christensen J., et al. UTX and JMJD3 Are Histone H3K27 Demethylases Involved in HOX Gene Regulation and Development. Nature 2007, 449, 731–734. - PubMed
-
- Lee M. G., Villa R., Trojer P., et al. Demethylation of H3K27 Regulates Polycomb Recruitment and H2A Ubiquitination. Science 2007, 318, 447–450. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

