QTLs associated with dry matter intake, metabolic mid-test weight, growth and feed efficiency have little overlap across 4 beef cattle studies
- PMID: 25410110
- PMCID: PMC4253998
- DOI: 10.1186/1471-2164-15-1004
QTLs associated with dry matter intake, metabolic mid-test weight, growth and feed efficiency have little overlap across 4 beef cattle studies
Abstract
Background: The identification of genetic markers associated with complex traits that are expensive to record such as feed intake or feed efficiency would allow these traits to be included in selection programs. To identify large-effect QTL, we performed a series of genome-wide association studies and functional analyses using 50 K and 770 K SNP genotypes scored in 5,133 animals from 4 independent beef cattle populations (Cycle VII, Angus, Hereford and Simmental×Angus) with phenotypes for average daily gain, dry matter intake, metabolic mid-test body weight and residual feed intake.
Results: A total of 5, 6, 11 and 10 significant QTL (defined as 1-Mb genome windows with Bonferroni-corrected P-value<0.05) were identified for average daily gain, dry matter intake, metabolic mid-test body weight and residual feed intake, respectively. The identified QTL were population-specific and had little overlap across the 4 populations. The pleiotropic or closely linked QTL on BTA 7 at 23 Mb identified in the Angus population harbours a promising candidate gene ACSL6 (acyl-CoA synthetase long-chain family member 6), and was the largest effect QTL associated with dry matter intake and mid-test body weight explaining 10.39% and 14.25% of the additive genetic variance, respectively. Pleiotropic or closely linked QTL associated with average daily gain and mid-test body weight were detected on BTA 6 at 38 Mb and BTA 7 at 93 Mb confirming previous reports. No QTL for residual feed intake explained more than 2.5% of the additive genetic variance in any population. Marker-based estimates of heritability ranged from 0.21 to 0.49 for residual feed intake across the 4 populations.
Conclusions: This GWAS study, which is the largest performed for feed efficiency and its component traits in beef cattle to date, identified several large-effect QTL that cumulatively explained a significant percentage of additive genetic variance within each population. Differences in the QTL identified among the different populations may be due to differences in power to detect QTL, environmental variation, or differences in the genetic architecture of trait variation among breeds. These results enhance our understanding of the biology of growth, feed intake and utilisation in beef cattle.
Figures
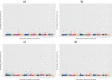
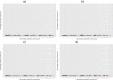
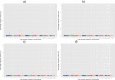


References
-
- Koch RM, Swiger LA, Chambers D, Gregory KE. Efficiency of feed use in beef cattle. J Anim Sci. 1963;22:486–494.
-
- Dickerson GE, Kunzi N, Cundiff LV, Koch RM, Arthaud VH, Gregory KE. Selection criteria for efficient beef production. J Anim Sci. 1974;39:659–673.
-
- Archer JA, Richardson EC, Herd RM, Arthur PF. Potential for selection to improve efficiency of feed use in beef cattle: a review. Austr J Agric Res. 1999;50:147–162. doi: 10.1071/A98075. - DOI
-
- Kennedy BW, van der Werf JH, Meuwissen TH. Genetic and statistical properties of residual feed intake. J Anim Sci. 1993;71:3239–3250. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

