Dissecting noncoding and pathogen RNA-protein interactomes
- PMID: 25411354
- PMCID: PMC4274633
- DOI: 10.1261/rna.047803.114
Dissecting noncoding and pathogen RNA-protein interactomes
Abstract
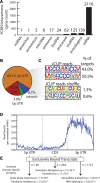
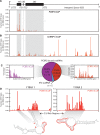
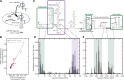
RNA-protein interactions are central to biological regulation. Cross-linking immunoprecipitation (CLIP)-seq is a powerful tool for genome-wide interrogation of RNA-protein interactomes, but current CLIP methods are limited by challenging biochemical steps and fail to detect many classes of noncoding and nonhuman RNAs. Here we present FAST-iCLIP, an integrated pipeline with improved CLIP biochemistry and an automated informatic pipeline for comprehensive analysis across protein coding, noncoding, repetitive, retroviral, and nonhuman transcriptomes. FAST-iCLIP of Poly-C binding protein 2 (PCBP2) showed that PCBP2-bound CU-rich motifs in different topologies to recognize mRNAs and noncoding RNAs with distinct biological functions. FAST-iCLIP of PCBP2 in hepatitis C virus-infected cells enabled a joint analysis of the PCBP2 interactome with host and viral RNAs and their interplay. These results show that FAST-iCLIP can be used to rapidly discover and decipher mechanisms of RNA-protein recognition across the diversity of human and pathogen RNAs.
Keywords: RNA–protein interactions; genomics; noncoding RNA; virology.
© 2014 Flynn et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

References
-
- Du Z, Lee JK, Tjhen R, Li S, Pan H, Stroud RM, James TL 2005. Crystal structure of the first KH domain of human poly(C)-binding protein-2 in complex with a C-rich strand of human telomeric DNA at 1.7 Å. J Biol Chem 280: 38823–38830. - PubMed
-
- Fraser CS, Doudna JA 2006. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol 5: 29–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
