Wavelet-based peak detection and a new charge inference procedure for MS/MS implemented in ProteoWizard's msConvert
- PMID: 25411686
- PMCID: PMC4324452
- DOI: 10.1021/pr500886y
Wavelet-based peak detection and a new charge inference procedure for MS/MS implemented in ProteoWizard's msConvert
Abstract
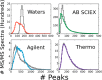
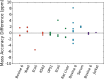
We report the implementation of high-quality signal processing algorithms into ProteoWizard, an efficient, open-source software package designed for analyzing proteomics tandem mass spectrometry data. Specifically, a new wavelet-based peak-picker (CantWaiT) and a precursor charge determination algorithm (Turbocharger) have been implemented. These additions into ProteoWizard provide universal tools that are independent of vendor platform for tandem mass spectrometry analyses and have particular utility for intralaboratory studies requiring the advantages of different platforms convergent on a particular workflow or for interlaboratory investigations spanning multiple platforms. We compared results from these tools to those obtained using vendor and commercial software, finding that in all cases our algorithms resulted in a comparable number of identified peptides for simple and complex samples measured on Waters, Agilent, and AB SCIEX quadrupole time-of-flight and Thermo Q-Exactive mass spectrometers. The mass accuracy of matched precursor ions also compared favorably with vendor and commercial tools. Additionally, typical analysis runtimes (∼1-100 ms per MS/MS spectrum) were short enough to enable the practical use of these high-quality signal processing tools for large clinical and research data sets.
Keywords: Continuous wavelet transformation; deisotoping; mass spectrometry; open-source software; peak-picking; precursor charge determination; signal deconvolution.
Figures


References
-
- Nesvizhskii A. I.; Vitek O.; Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods. 2007, 4, 787–797. - PubMed
-
- Bantscheff M.; Lemeer S.; Savitski M. M.; Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 2012, 404, 939–65. - PubMed
-
- Cox J.; Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

