Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011
- PMID: 25411691
- PMCID: PMC4518193
- DOI: 10.2807/1560-7917.es2014.19.45.20954
Whole genome sequencing reveals potential spread of Clostridium difficile between humans and farm animals in the Netherlands, 2002 to 2011
Abstract
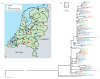
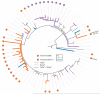
Farm animals are a potential reservoir for human Clostridium difficile infection (CDI), particularly PCR ribotype 078 which is frequently found in animals and humans. Here, whole genome single-nucleotide polymorphism (SNP) analysis was used to study the evolutionary relatedness of C. difficile 078 isolated from humans and animals on Dutch pig farms. All sequenced genomes were surveyed for potential antimicrobial resistance determinants and linked to an antimicrobial resistance phenotype. We sequenced the whole genome of 65 C. difficile 078 isolates collected between 2002 and 2011 from pigs (n = 19), asymptomatic farmers (n = 15) and hospitalised patients (n = 31) in the Netherlands. The collection included 12 pairs of human and pig isolates from 2011 collected at 12 different pig farms. A mutation rate of 1.1 SNPs per genome per year was determined for C. difficile 078. Importantly, we demonstrate that farmers and pigs were colonised with identical (no SNP differences) and nearly identical (less than two SNP differences) C. difficile clones. Identical tetracycline and streptomycin resistance determinants were present in human and animal C. difficile 078 isolates. Our observation that farmers and pigs share identical C. difficile strains suggests transmission between these populations, although we cannot exclude the possibility of transmission from a common environmental source.
Figures

References
-
- Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. 1994;330(4):257–62. http://dx.doi.org/10.1056/NEJM199401273300406 PMID:8043060. - DOI - PubMed
-
- Bartlett JG, Gerding DN. Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis. 2008;46((s1) Suppl 1):S12–8. http://dx.doi.org/10.1086/521863 PMID:18177217. - DOI - PubMed
-
- Dial S, Delaney JA, Barkun AN, Suissa S. Use of gastric acid-suppressive agents and the risk of community-acquired Clostridium difficile-associated disease. JAMA. 2005;294(23):2989–95. http://dx.doi.org/10.1001/jama.294.23.2989 PMID:16414946. - DOI - PubMed
-
- Dial S, Delaney JA, Schneider V, Suissa S. Proton pump inhibitor use and risk of community-acquired Clostridium difficile-associated disease defined by prescription for oral vancomycin therapy. CMAJ. 2006;175(7):745–8. http://dx.doi.org/10.1503/cmaj.060284 PMID:17001054. - DOI - PMC - PubMed
-
- Wilcox MH, Mooney L, Bendall R, Settle CD, Fawley WN. A case-control study of community-associated Clostridium difficile infection. J Antimicrob Chemother. 2008;62(2):388–96. http://dx.doi.org/10.1093/jac/dkn163 PMID:18434341. - DOI - PubMed

