High prevalence of co-infection with multiple Torque teno sus virus species in Italian pig herds
- PMID: 25411972
- PMCID: PMC4239083
- DOI: 10.1371/journal.pone.0113720
High prevalence of co-infection with multiple Torque teno sus virus species in Italian pig herds
Abstract
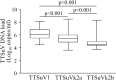
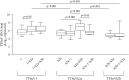
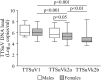
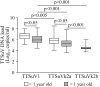
Torque teno viruses (TTVs) are a large group of vertebrate-infecting small viruses with circular single-stranded DNA, classified in the Anelloviridae family. In swine, two genetically distinct species, Torque teno sus virus 1a (TTSuV1a) and 1b (TTSuV1b) are currently grouped into the genus Iotatorquevirus. More recently, a novel Torque teno sus virus species, named Torque teno sus virus k2b (TTSuVk2b), has been included with Torque teno sus virus k2a (TTSuVk2a) into the genus Kappatorquevirus. In the present study, TTSuV1 (TTSuV1a and TTSuV1b), TTSuVk2a and TTSuVk2b prevalence was evaluated in 721 serum samples of healthy pigs from Sardinian farms, insular Italy. This is the largest study to date on the presence of TTSuV in healthy pigs in Italy. The global prevalence of infection was 83.2% (600/721), being 62.3% (449/721), 60.6% (437/721), and 11.5% (83/721) the prevalence of TTSuV1, TTSuVk2a and TTSuVk2b, respectively. The rate of co-infection with two and/or three species was also calculated, and data show that co-infections were significantly more frequent than infections with single species, and that TTSuV1+TTSuVk2a double infection was the prevalent combination (35.4%). Quantitative results obtained using species-specific real time-qPCR evidenced the highest mean levels of viremia in the TTSuV1 subgroup, and the lowest in the TTSuVk2b subgroup. Interestingly, multiple infections with distinct TTSuV species seemed to significantly affect the DNA load and specifically, data highlighted that double infection with TTSuVk2a increased the viral titers of TTSuV1, likewise the co-infection with TTSuVk2b increased the titers of TTSuVk2a.
Conflict of interest statement
Figures
References
-
- Allan GM, Mc Neilly F, Meehan BM, Kennedy S, Mackie DP, et al. (1999) Isolation and characterization of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet Microbiol 66: 115–123. - PubMed
-
- Nishizawa T, Okamoto H, Konishi K, Yoshizawa H, Miyakawa Y, et al. (1997) A novel DNA virus (TTV) associated with elevated transaminase levels in posttransfusion hepatitis of unknown etiology. Biochem Biophys Res Commun 241: 92–97. - PubMed
-
- Okamoto H, Takahashi M, Nishizawa T, Tawara A, Fukai K, et al. (2002) Genomic characterization of TT viruses (TTVs) in pigs, cats and dogs and their relatedness with species-specific TTVs in primates and tupaias. J Gen Virol 83: 1291–1297. - PubMed
-
- Niel C, Diniz-Mendes L, Devalle S (2005) Rolling-circle amplification of Torque teno virus (TTV) complete genomes from human and swine sera and identification of a novel swine TTV genogroup. J Gen Virol 86: 1343–1347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

