Combination simvastatin and metformin induces G1-phase cell cycle arrest and Ripk1- and Ripk3-dependent necrosis in C4-2B osseous metastatic castration-resistant prostate cancer cells
- PMID: 25412314
- PMCID: PMC4260755
- DOI: 10.1038/cddis.2014.500
Combination simvastatin and metformin induces G1-phase cell cycle arrest and Ripk1- and Ripk3-dependent necrosis in C4-2B osseous metastatic castration-resistant prostate cancer cells
Abstract
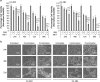
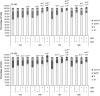
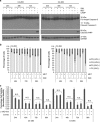
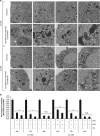
Castration-resistant prostate cancer (CRPC) cells acquire resistance to chemotherapy and apoptosis, in part, due to enhanced aerobic glycolysis and biomass production, known as the Warburg effect. We previously demonstrated that combination simvastatin (SIM) and metformin (MET) ameliorates critical Warburg effect-related metabolic aberrations of C4-2B cells, synergistically and significantly decreases CRPC cell viability and metastatic properties, with minimal effect on normal prostate epithelial cells, and inhibits primary prostate tumor growth, metastasis, and biochemical failure in an orthotopic model of metastatic CRPC, more effectively than docetaxel chemotherapy. Several modes of cell death activated by individual treatment of SIM or MET have been reported; however, the cell death process induced by combination SIM and MET treatment in metastatic CRPC cells remains unknown. This must be determined prior to advancing combination SIM and MET to clinical trial for metastatic CRPC. Treatment of C4-2B cells with combination 4 μM SIM and 2 mM MET (SIM+MET) led to significant G1-phase cell cycle arrest and decrease in the percentage of DNA-replicating cells in the S-phase by 24 h; arrest was sustained throughout the 96-h treatment. SIM+MET treatment led to enhanced autophagic flux in C4-2B cells by 72-96 h, ascertained by increased LC3B-II (further enhanced with lysosomal inhibitor chloroquine) and reduced Sequestosome-1 protein expression, significantly increased percentage of acidic vesicular organelle-positive cells, and increased autophagic structure accumulation assessed by transmission electron microscopy. Chloroquine, however, could not rescue CRPC cell viability, eliminating autophagic cell death; rather, autophagy was upregulated by C4-2B cells in attempt to withstand chemotherapy. Instead, SIM+MET treatment led to Ripk1- and Ripk3-dependent necrosis by 48-96 h, determined by propidium iodide-Annexin V flow cytometry, increase in Ripk1 and Ripk3 protein expression, necrosome formation, HMGB-1 extracellular release, and necrotic induction and viability rescue with necrostatin-1 and Ripk3-targeting siRNA. The necrosis-inducing capacity of SIM+MET may make these drugs a highly-effective treatment for apoptosis- and chemotherapy-resistant metastatic CRPC cells.
Figures





References
-
- American Cancer Society. Cancer Facts & Figures 2014. American Cancer Society: Atlanta, GA, USA; 2014.
-
- Cookson MS, Roth BJ, Dahm P, Engstrom C, Freedland SJ, Hussain M, et al. Castration-resistant prostate cancer: AUA Guideline. J Urol. 2013;190:429–438. - PubMed
-
- Loblaw DA, Walker-Dilks C, Winquist E, Hotte SJ, Genitourinary Cancer Disease Site Group of Cancer Care Ontario's Program in Evidence-Based Care Systemic therapy in men with metastatic castration-resistant prostate cancer: a systematic review. Clin Oncol (R Coll Radiol) 2013;25:406–430. - PubMed
-
- Tannock IF, de Wit R, Berry WR, Horti J, Pluzanska A, Chi KN, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

