Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing
- PMID: 25412400
- PMCID: PMC4572572
- DOI: 10.1038/gim.2014.172
Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing
Abstract
Purpose: Next-generation sequencing-based methods are being adopted broadly for genetic diagnostic testing, but the performance characteristics of these techniques with regard to test accuracy and reproducibility have not been fully defined.
Methods: We developed a targeted enrichment and next-generation sequencing approach for genetic diagnostic testing of patients with inherited eye disorders, including inherited retinal degenerations, optic atrophy, and glaucoma. In preparation for providing this genetic eye disease (GEDi) test on a CLIA-certified basis, we performed experiments to measure the sensitivity, specificity, and reproducibility, as well as the clinical sensitivity, of the test.
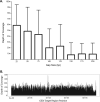
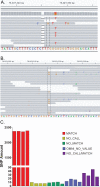
Results: The GEDi test is highly reproducible and accurate, with sensitivity and specificity of 97.9 and 100%, respectively, for single-nucleotide variant detection. The sensitivity for variant detection was notably better than the 88.3% achieved by whole-exome sequencing using the same metrics, because of better coverage of targeted genes in the GEDi test as compared with a commercially available exome capture set. Prospective testing of 192 patients with inherited retinal degenerations indicated that the clinical sensitivity of the GEDi test is high, with a diagnostic rate of 51%.
Conclusion: Based on quantified performance metrics, the data suggest that selective targeted enrichment is preferable to whole-exome sequencing for genetic diagnostic testing.
Figures
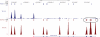
References
-
- Sommen M, Van Camp G. Genetic diagnostics of early childhood hearing loss: better testing with next-generation DNA sequencing. B-Ent. 2013;(Suppl 21):51–56. - PubMed
-
- Pugh TJ, Kelly MA, Gowrisankar S, et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genetics In Medicine. 2014 Feb 6; - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

