Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha
- PMID: 25412462
- PMCID: PMC4238957
- DOI: 10.1371/journal.pgen.1004796
Inversion of the chromosomal region between two mating type loci switches the mating type in Hansenula polymorpha
Abstract
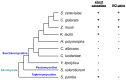
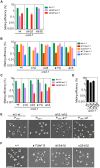
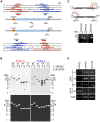
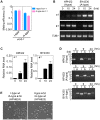
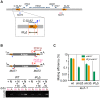
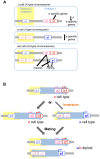
Yeast mating type is determined by the genotype at the mating type locus (MAT). In homothallic (self-fertile) Saccharomycotina such as Saccharomyces cerevisiae and Kluveromyces lactis, high-efficiency switching between a and α mating types enables mating. Two silent mating type cassettes, in addition to an active MAT locus, are essential components of the mating type switching mechanism. In this study, we investigated the structure and functions of mating type genes in H. polymorpha (also designated as Ogataea polymorpha). The H. polymorpha genome was found to harbor two MAT loci, MAT1 and MAT2, that are ∼18 kb apart on the same chromosome. MAT1-encoded α1 specifies α cell identity, whereas none of the mating type genes were required for a identity and mating. MAT1-encoded α2 and MAT2-encoded a1 were, however, essential for meiosis. When present in the location next to SLA2 and SUI1 genes, MAT1 or MAT2 was transcriptionally active, while the other was repressed. An inversion of the MAT intervening region was induced by nutrient limitation, resulting in the swapping of the chromosomal locations of two MAT loci, and hence switching of mating type identity. Inversion-deficient mutants exhibited severe defects only in mating with each other, suggesting that this inversion is the mechanism of mating type switching and homothallism. This chromosomal inversion-based mechanism represents a novel form of mating type switching that requires only two MAT loci.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Regulation of mating type switching by the mating type genes and RME1 in Ogataea polymorpha.Sci Rep. 2017 Nov 24;7(1):16318. doi: 10.1038/s41598-017-16284-7. Sci Rep. 2017. PMID: 29176579 Free PMC article.
-
The Sclerotinia sclerotiorum mating type locus (MAT) contains a 3.6-kb region that is inverted in every meiotic generation.PLoS One. 2013;8(2):e56895. doi: 10.1371/journal.pone.0056895. Epub 2013 Feb 15. PLoS One. 2013. PMID: 23457637 Free PMC article.
-
Mating-type switching by chromosomal inversion in methylotrophic yeasts suggests an origin for the three-locus Saccharomyces cerevisiae system.Proc Natl Acad Sci U S A. 2014 Nov 11;111(45):E4851-8. doi: 10.1073/pnas.1416014111. Epub 2014 Oct 27. Proc Natl Acad Sci U S A. 2014. PMID: 25349420 Free PMC article.
-
An Evolutionary Perspective on Yeast Mating-Type Switching.Genetics. 2017 May;206(1):9-32. doi: 10.1534/genetics.117.202036. Genetics. 2017. PMID: 28476860 Free PMC article. Review.
-
Evolution of Mating in the Saccharomycotina.Annu Rev Microbiol. 2017 Sep 8;71:197-214. doi: 10.1146/annurev-micro-090816-093403. Epub 2017 Jun 28. Annu Rev Microbiol. 2017. PMID: 28657889 Review.
Cited by
-
Pondering Mating: Pneumocystis jirovecii, the Human Lung Pathogen, Selfs without Mating Type Switching, in Contrast to Its Close Relative Schizosaccharomyces pombe.mBio. 2015 May 5;6(3):e00583-15. doi: 10.1128/mBio.00583-15. mBio. 2015. PMID: 25944864 Free PMC article. No abstract available.
-
Draft genome of the Ogataea polymorpha type strain CBS 4732.Microbiol Resour Announc. 2023 Sep 19;12(9):e0061122. doi: 10.1128/MRA.00611-22. Epub 2023 Aug 2. Microbiol Resour Announc. 2023. PMID: 37530527 Free PMC article.
-
From Two to One: Unipolar Sexual Reproduction.Fungal Biol Rev. 2015 Dec 1;29(3-4):118-125. doi: 10.1016/j.fbr.2015.06.002. Epub 2015 Jul 10. Fungal Biol Rev. 2015. PMID: 26744600 Free PMC article.
-
Genome Diversity and Evolution in the Budding Yeasts (Saccharomycotina).Genetics. 2017 Jun;206(2):717-750. doi: 10.1534/genetics.116.199216. Genetics. 2017. PMID: 28592505 Free PMC article. Review.
-
Strains and approaches for genetic crosses in the oleaginous yeast Lipomyces starkeyi.Yeast. 2021 Dec;38(12):625-633. doi: 10.1002/yea.3671. Epub 2021 Oct 15. Yeast. 2021. PMID: 34596906 Free PMC article.
References
-
- Herskowitz I (1989) A regulatory hierarchy for cell specialization in yeast. Nature 342: 749–757. - PubMed
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

