Divergent paths for the selection of immunodominant epitopes from distinct antigenic sources
- PMID: 25413013
- PMCID: PMC4241505
- DOI: 10.1038/ncomms6369
Divergent paths for the selection of immunodominant epitopes from distinct antigenic sources
Abstract
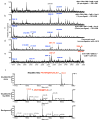
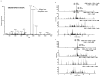
Immunodominant epitopes are few selected epitopes from complex antigens that initiate T-cell responses. Here to provide further insights into this process, we use a reductionist cell-free antigen-processing system composed of defined components. We use the system to characterize steps in antigen processing of pathogen-derived proteins or autoantigens and we find distinct paths for peptide processing and selection. Autoantigen-derived immunodominant epitopes are resistant to digestion by cathepsins, whereas pathogen-derived epitopes are sensitive. Sensitivity to cathepsins enforces capture of pathogen-derived epitopes by major histocompatibility complex class II (MHC class II) before processing, and resistance to HLA-DM-mediated-dissociation preserves the longevity of those epitopes. We show that immunodominance is established by higher relative abundance of the selected epitopes, which survive cathepsin digestion either by binding to MHC class II and resisting DM-mediated-dissociation, or being chemically resistant to cathepsins degradation. Non-dominant epitopes are sensitive to both DM and cathepsins and are destroyed.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
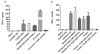






Similar articles
-
A reductionist cell-free major histocompatibility complex class II antigen processing system identifies immunodominant epitopes.Nat Med. 2010 Nov;16(11):1333-40. doi: 10.1038/nm.2248. Epub 2010 Oct 31. Nat Med. 2010. PMID: 21037588 Free PMC article.
-
Antigen-presenting cells and the selection of immunodominant epitopes.Crit Rev Immunol. 1997;17(5-6):411-7. Crit Rev Immunol. 1997. PMID: 9419428 Review.
-
Exogenous antigens bind MHC class II first, and are processed by cathepsins later.Mol Immunol. 2015 Dec;68(2 Pt A):81-4. doi: 10.1016/j.molimm.2015.07.018. Epub 2015 Aug 5. Mol Immunol. 2015. PMID: 26254987 Free PMC article. Review.
-
Characterizing immunodominant and protective influenza hemagglutinin epitopes by functional activity and relative binding to major histocompatibility complex class II sites.Eur J Immunol. 1997 Dec;27(12):3105-14. doi: 10.1002/eji.1830271205. Eur J Immunol. 1997. PMID: 9464794
-
Selection of immunodominant epitopes during antigen processing is hierarchical.Mol Immunol. 2019 Sep;113:115-119. doi: 10.1016/j.molimm.2018.08.011. Epub 2018 Aug 24. Mol Immunol. 2019. PMID: 30146122 Free PMC article. Review.
Cited by
-
Proper development of long-lived memory CD4 T cells requires HLA-DO function.Front Immunol. 2023 Oct 16;14:1277609. doi: 10.3389/fimmu.2023.1277609. eCollection 2023. Front Immunol. 2023. PMID: 37908352 Free PMC article.
-
A cell-free antigen processing system informs HIV-1 epitope selection and vaccine design.J Exp Med. 2023 Jul 3;220(7):e20221654. doi: 10.1084/jem.20221654. Epub 2023 Apr 14. J Exp Med. 2023. PMID: 37058141 Free PMC article.
-
Epitope Selection for HLA-DQ2 Presentation: Implications for Celiac Disease and Viral Defense.J Immunol. 2019 May 1;202(9):2558-2569. doi: 10.4049/jimmunol.1801454. Epub 2019 Mar 29. J Immunol. 2019. PMID: 30926644 Free PMC article.
-
Human Cysteine Cathepsins Degrade Immunoglobulin G In Vitro in a Predictable Manner.Int J Mol Sci. 2019 Sep 29;20(19):4843. doi: 10.3390/ijms20194843. Int J Mol Sci. 2019. PMID: 31569504 Free PMC article.
-
Human leukocyte Antigen-DM polymorphisms in autoimmune diseases.Open Biol. 2016 Aug;6(8):160165. doi: 10.1098/rsob.160165. Open Biol. 2016. PMID: 27534821 Free PMC article. Review.
References
-
- Sercarz EE, Maverakis E. Mhc-guided processing: binding of large antigen fragments. Nat Rev Immunol. 2003;3:621–629. - PubMed
-
- Dai G, Carmicle S, Steede NK, Landry SJ. Structural basis for helper T-cell and antibody epitope immunodominance in bacteriophage T4 Hsp10. Role of disordered loops. J Biol Chem. 2002;277:161–168. - PubMed
-
- Maric M, et al. Defective antigen processing in GILT-free mice. Science. 2001;294:1361–1365. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

