SR proteins control a complex network of RNA-processing events
- PMID: 25414008
- PMCID: PMC4274639
- DOI: 10.1261/rna.043893.113
SR proteins control a complex network of RNA-processing events
Abstract
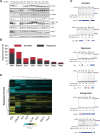
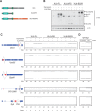
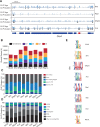
SR proteins are a well-conserved class of RNA-binding proteins that are essential for regulation of splice-site selection, and have also been implicated as key regulators during other stages of RNA metabolism. For many SR proteins, the complexity of the RNA targets and specificity of RNA-binding location are poorly understood. It is also unclear if general rules governing SR protein alternative pre-mRNA splicing (AS) regulation uncovered for individual SR proteins on few model genes, apply to the activity of all SR proteins on endogenous targets. Using RNA-seq, we characterize the global AS regulation of the eight Drosophila SR protein family members. We find that a majority of AS events are regulated by multiple SR proteins, and that all SR proteins can promote exon inclusion, but also exon skipping. Most coregulated targets exhibit cooperative regulation, but some AS events are antagonistically regulated. Additionally, we found that SR protein levels can affect alternative promoter choices and polyadenylation site selection, as well as overall transcript levels. Cross-linking and immunoprecipitation coupled with high-throughput sequencing (iCLIP-seq), reveals that SR proteins bind a distinct and functionally diverse class of RNAs, which includes several classes of noncoding RNAs, uncovering possible novel functions of the SR protein family. Finally, we find that SR proteins exhibit positional RNA binding around regulated AS events. Therefore, regulation of AS by the SR proteins is the result of combinatorial regulation by multiple SR protein family members on most endogenous targets, and SR proteins have a broader role in integrating multiple layers of gene expression regulation.
Keywords: RNA processing; SR proteins; alternative pre-mRNA splicing; regulation of gene expression.
© 2014 Bradley et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures




References
-
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
