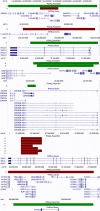
DNA copy number variants of known glaucoma genes in relation to primary open-angle glaucoma
- PMID: 25414181
- PMCID: PMC4271633
- DOI: 10.1167/iovs.14-15712
DNA copy number variants of known glaucoma genes in relation to primary open-angle glaucoma
Abstract
Purpose: We examined the role of DNA copy number variants (CNVs) of known glaucoma genes in relation to primary open angle glaucoma (POAG).
Methods: Our study included DNA samples from two studies (NEIGHBOR and GLAUGEN). All the samples were genotyped with the Illumina Human660W_Quad_v1 BeadChip. After removing non-blood-derived and amplified DNA samples, we applied quality control steps based on the mean Log R Ratio and the mean B allele frequency. Subsequently, data from 3057 DNA samples (1599 cases and 1458 controls) were analyzed with PennCNV software. We defined CNVs as those ≥5 kilobases (kb) in size and interrogated by ≥5 consecutive probes. We further limited our investigation to CNVs in known POAG-related genes, including CDKN2B-AS1, TMCO1, SIX1/SIX6, CAV1/CAV2, the LRP12-ZFPM2 region, GAS7, ATOH7, FNDC3B, CYP1B1, MYOC, OPTN, WDR36, SRBD1, TBK1, and GALC.
Results: Genomic duplications of CDKN2B-AS1 and TMCO1 were each found in a single case. Two cases carried duplications in the GAS7 region. Genomic deletions of SIX6 and ATOH7 were each identified in one case. One case carried a TBK1 deletion and another case carried a TBK1 duplication. No controls had duplications or deletions in these six genes. A single control had a duplication in the MYOC region. Deletions of GALC were observed in five cases and two controls.
Conclusions: The CNV analysis of a large set of cases and controls revealed the presence of rare CNVs in known POAG susceptibility genes. Our data suggest that these rare CNVs may contribute to POAG pathogenesis and merit functional evaluation.
Keywords: DNA copy number variants; GAS7; POAG; SIX6; genetics.
Copyright 2014 The Association for Research in Vision and Ophthalmology, Inc.
Figures
Similar articles
-
Genetic Variants Associated With Different Risks for High Tension Glaucoma and Normal Tension Glaucoma in a Chinese Population.Invest Ophthalmol Vis Sci. 2015 Apr;56(4):2595-600. doi: 10.1167/iovs.14-16269. Invest Ophthalmol Vis Sci. 2015. PMID: 25711633
-
Genetic analysis of primary open-angle glaucoma-related risk alleles in a Korean population: the GLAU-GENDISK study.Br J Ophthalmol. 2021 Sep;105(9):1307-1312. doi: 10.1136/bjophthalmol-2020-316089. Epub 2020 Sep 15. Br J Ophthalmol. 2021. PMID: 32933932
-
Association of known common genetic variants with primary open angle, primary angle closure, and pseudoexfoliation glaucoma in Pakistani cohorts.Mol Vis. 2014 Nov 4;20:1471-9. eCollection 2014. Mol Vis. 2014. PMID: 25489222 Free PMC article.
-
Molecular Genetics of Glaucoma: Subtype and Ethnicity Considerations.Genes (Basel). 2020 Dec 31;12(1):55. doi: 10.3390/genes12010055. Genes (Basel). 2020. PMID: 33396423 Free PMC article. Review.
-
An Updated Review on the Genetics of Primary Open Angle Glaucoma.Int J Mol Sci. 2015 Dec 4;16(12):28886-911. doi: 10.3390/ijms161226135. Int J Mol Sci. 2015. PMID: 26690118 Free PMC article. Review.
Cited by
-
SQSTM1 Mutations and Glaucoma.PLoS One. 2016 Jun 8;11(6):e0156001. doi: 10.1371/journal.pone.0156001. eCollection 2016. PLoS One. 2016. PMID: 27275741 Free PMC article.
-
The Role of Mitophagy in Glaucomatous Neurodegeneration.Cells. 2023 Jul 30;12(15):1969. doi: 10.3390/cells12151969. Cells. 2023. PMID: 37566048 Free PMC article. Review.
-
Candidate SNP Markers Significantly Altering the Affinity of the TATA-Binding Protein for the Promoters of Human Genes Associated with Primary Open-Angle Glaucoma.Int J Mol Sci. 2024 Nov 28;25(23):12802. doi: 10.3390/ijms252312802. Int J Mol Sci. 2024. PMID: 39684516 Free PMC article.
-
Genetic association between CDKN2B-AS1 polymorphisms and the susceptibility of primary open-angle glaucoma (POAG): a meta-analysis from 21,775 subjects.Ir J Med Sci. 2022 Oct;191(5):2385-2392. doi: 10.1007/s11845-021-02794-x. Epub 2021 Oct 14. Ir J Med Sci. 2022. PMID: 34648117 Free PMC article.
-
Transgenic TBK1 mice have features of normal tension glaucoma.Hum Mol Genet. 2017 Jan 1;26(1):124-132. doi: 10.1093/hmg/ddw372. Hum Mol Genet. 2017. PMID: 28025332 Free PMC article.
References
-
- Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014; 121: 2081–2090. - PubMed
-
- Liu Y, Allingham RR. Genetics of glaucoma. Encyclopedia of Life Sciences (ELS). Chichester, UK: John Wiley & Sons Ltd; 2010.
-
- Allingham RR, Shields MB. Shields' Textbook of Glaucoma. 6th ed. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health; 2011.
Publication types
MeSH terms
Substances
Grants and funding
- R01 EY011671/EY/NEI NIH HHS/United States
- P30-EY005722/EY/NEI NIH HHS/United States
- R01EY022305/EY/NEI NIH HHS/United States
- T32 EY021453/EY/NEI NIH HHS/United States
- EY023512/EY/NEI NIH HHS/United States
- R01 EY015473/EY/NEI NIH HHS/United States
- EY009149/EY/NEI NIH HHS/United States
- R01 EY022306/EY/NEI NIH HHS/United States
- R01 EY013315/EY/NEI NIH HHS/United States
- EY144428/EY/NEI NIH HHS/United States
- U01 HG004728/HG/NHGRI NIH HHS/United States
- T32EY021453/EY/NEI NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- P30 EY005722/EY/NEI NIH HHS/United States
- EY012118/EY/NEI NIH HHS/United States
- UL1 TR000427/TR/NCATS NIH HHS/United States
- R01 EY011008/EY/NEI NIH HHS/United States
- P01 CA087969/CA/NCI NIH HHS/United States
- EY18660/EY/NEI NIH HHS/United States
- EY015473/EY/NEI NIH HHS/United States
- EY015872/EY/NEI NIH HHS/United States
- EY010886/EY/NEI NIH HHS/United States
- CA87969/CA/NCI NIH HHS/United States
- EY009847/EY/NEI NIH HHS/United States
- U01 HG004424/HG/NHGRI NIH HHS/United States
- R01 HL073389/HL/NHLBI NIH HHS/United States
- EY011671/EY/NEI NIH HHS/United States
- CA49449/CA/NCI NIH HHS/United States
- EY13315/EY/NEI NIH HHS/United States
- HG005259-01/HG/NHGRI NIH HHS/United States
- EY015682/EY/NEI NIH HHS/United States
- R01 EY023242/EY/NEI NIH HHS/United States
- EY019126/EY/NEI NIH HHS/United States
- R01 EY012118/EY/NEI NIH HHS/United States
- R01 EY015543/EY/NEI NIH HHS/United States
- R56 EY011671/EY/NEI NIH HHS/United States
- R01EY023242/EY/NEI NIH HHS/United States
- R01 EY008208/EY/NEI NIH HHS/United States
- HG004608/HG/NHGRI NIH HHS/United States
- HG004728/HG/NHGRI NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- EY013178/EY/NEI NIH HHS/United States
- R01 EY013178/EY/NEI NIH HHS/United States
- EY09611/EY/NEI NIH HHS/United States
- EY006827/EY/NEI NIH HHS/United States
- R01 EY019126/EY/NEI NIH HHS/United States
- R03 EY015682/EY/NEI NIH HHS/United States
- R01 EY022305/EY/NEI NIH HHS/United States
- HG004446/HG/NHGRI NIH HHS/United States
- HL35464/HL/NHLBI NIH HHS/United States
- R01 EY009611/EY/NEI NIH HHS/United States
- R01 EY011405/EY/NEI NIH HHS/United States
- EY011008/EY/NEI NIH HHS/United States
- UM1 CA167552/CA/NCI NIH HHS/United States
- R01 EY009580/EY/NEI NIH HHS/United States
- R01 HL035464/HL/NHLBI NIH HHS/United States
- U10 EY012118/EY/NEI NIH HHS/United States
- R01 CA049449/CA/NCI NIH HHS/United States
- P30 EY014104/EY/NEI NIH HHS/United States
- R01 EY015872/EY/NEI NIH HHS/United States
- R01 EY009847/EY/NEI NIH HHS/United States
- R01 EY010886/EY/NEI NIH HHS/United States
- U01 CA049449/CA/NCI NIH HHS/United States
- HL073389/HL/NHLBI NIH HHS/United States
- EY09580/EY/NEI NIH HHS/United States
- HG004424/HG/NHGRI NIH HHS/United States
- EY015543/EY/NEI NIH HHS/United States
- EY008208/EY/NEI NIH HHS/United States
- U01 HG004608/HG/NHGRI NIH HHS/United States
- R01 EY023512/EY/NEI NIH HHS/United States
- R01 EY018660/EY/NEI NIH HHS/United States
- EY144448/EY/NEI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous