MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor (MITF) mRNA is inhibited by coding region determinant-binding protein (CRD-BP)
- PMID: 25414259
- PMCID: PMC4281741
- DOI: 10.1074/jbc.M114.590158
MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor (MITF) mRNA is inhibited by coding region determinant-binding protein (CRD-BP)
Abstract
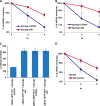
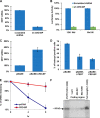
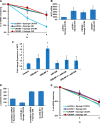
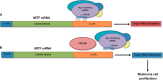
Alternative cleavage and polyadenylation generates multiple transcript variants producing mRNA isoforms with different length 3'-UTRs. Alternative cleavage and polyadenylation enables differential post-transcriptional regulation via the availability of different cis-acting elements in 3'-UTRs. Microphthalmia-associated transcription factor (MITF) is a master regulator of melanocyte development and melanogenesis. This central transcription factor is also implicated in melanoma development. Here, we show that melanoma cells favor the expression of MITF mRNA with a shorter 3'-UTR. We also establish that this isoform is regulated by a micro RNA (miRNA/miR), miR-340. miR-340 interacts with two of its target sites on the MITF 3'-UTR, causing mRNA degradation as well as decreased expression and activity of MITF. Conversely, the RNA-binding protein, coding region determinant-binding protein, was shown to be highly expressed in melanoma, directly binds to the 3'-UTR of MITF mRNA, and prevents the binding of miR-340 to its target sites, resulting in the stabilization of MITF transcripts, elevated expression, and transcriptional activity of MITF. This regulatory interplay between RNA-binding protein and miRNA highlights an important mechanism for the regulation of MITF in melanocytes and malignant melanomas.
Keywords: Alternative Cleavage and Polyadenylation; CRD-BP; IGF2BP1; MITF; Melanogenesis; Melanoma; MicroRNA (miRNA); Ribonuclear Protein (RNP); mRNA; mRNA Decay; miRNA.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Characterizing the Coding Region Determinant-Binding Protein (CRD-BP)-Microphthalmia-associated Transcription Factor (MITF) mRNA interaction.PLoS One. 2017 Feb 9;12(2):e0171196. doi: 10.1371/journal.pone.0171196. eCollection 2017. PLoS One. 2017. PMID: 28182633 Free PMC article.
-
MicroRNA-340-mediated degradation of microphthalmia-associated transcription factor mRNA is inhibited by the coding region determinant-binding protein.J Biol Chem. 2010 Jul 2;285(27):20532-40. doi: 10.1074/jbc.M110.109298. Epub 2010 May 3. J Biol Chem. 2010. Retraction in: J Biol Chem. 2014 Apr 25;289(17):11859. doi: 10.1074/jbc.A110.109298. PMID: 20439467 Free PMC article. Retracted.
-
MicroRNA-218 inhibits melanogenesis by directly suppressing microphthalmia-associated transcription factor expression.RNA Biol. 2014;11(6):732-41. doi: 10.4161/rna.28865. Epub 2014 Apr 24. RNA Biol. 2014. PMID: 24824743 Free PMC article.
-
[The Importance of MITF Signaling Pathway in the Regulation of Proliferation and Invasiveness of Malignant Melanoma].Klin Onkol. 2016 Fall;29(5):347-350. doi: 10.14735/amko2016347. Klin Onkol. 2016. PMID: 27739313 Review. Czech.
-
A big gene linked to small eyes encodes multiple Mitf isoforms: many promoters make light work.Pigment Cell Res. 1998 Dec;11(6):329-36. doi: 10.1111/j.1600-0749.1998.tb00491.x. Pigment Cell Res. 1998. PMID: 9870544 Review.
Cited by
-
Inhibition of the mRNA-Binding Protein IGF2BP1 Suppresses Proliferation and Sensitizes Neuroblastoma Cells to Chemotherapeutic Agents.Front Oncol. 2021 Mar 16;11:608816. doi: 10.3389/fonc.2021.608816. eCollection 2021. Front Oncol. 2021. PMID: 33796454 Free PMC article.
-
Unravelling Tumour Microenvironment in Melanoma at Single-Cell Level and Challenges to Checkpoint Immunotherapy.Genes (Basel). 2022 Sep 28;13(10):1757. doi: 10.3390/genes13101757. Genes (Basel). 2022. PMID: 36292642 Free PMC article. Review.
-
The regulatory impact of RNA-binding proteins on microRNA targeting.Nat Commun. 2021 Aug 20;12(1):5057. doi: 10.1038/s41467-021-25078-5. Nat Commun. 2021. PMID: 34417449 Free PMC article.
-
Characterizing the Coding Region Determinant-Binding Protein (CRD-BP)-Microphthalmia-associated Transcription Factor (MITF) mRNA interaction.PLoS One. 2017 Feb 9;12(2):e0171196. doi: 10.1371/journal.pone.0171196. eCollection 2017. PLoS One. 2017. PMID: 28182633 Free PMC article.
-
IGF2BP1, a Conserved Regulator of RNA Turnover in Cancer.Front Mol Biosci. 2021 Mar 22;8:632219. doi: 10.3389/fmolb.2021.632219. eCollection 2021. Front Mol Biosci. 2021. PMID: 33829040 Free PMC article.
References
-
- Hemesath T. J., Steingrímsson E., McGill G., Hansen M. J., Vaught J., Hodgkinson C. A., Arnheiter H., Copeland N. G., Jenkins N. A., Fisher D. E. (1994) Microphthalmia, a critical factor in melanocyte development, defines a discrete transcription factor family. Genes Dev. 8, 2770–2780 - PubMed
-
- Carreira S., Liu B., Goding C. R. (2000) The gene encoding the T-box factor Tbx2 is a target for the microphthalmia-associated transcription factor in melanocytes. J. Biol. Chem. 275, 21920–21927 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

