Araport: the Arabidopsis information portal
- PMID: 25414324
- PMCID: PMC4383980
- DOI: 10.1093/nar/gku1200
Araport: the Arabidopsis information portal
Abstract
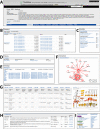
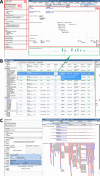
The Arabidopsis Information Portal (https://www.araport.org) is a new online resource for plant biology research. It houses the Arabidopsis thaliana genome sequence and associated annotation. It was conceived as a framework that allows the research community to develop and release 'modules' that integrate, analyze and visualize Arabidopsis data that may reside at remote sites. The current implementation provides an indexed database of core genomic information. These data are made available through feature-rich web applications that provide search, data mining, and genome browser functionality, and also by bulk download and web services. Araport uses software from the InterMine and JBrowse projects to expose curated data from TAIR, GO, BAR, EBI, UniProt, PubMed and EPIC CoGe. The site also hosts 'science apps,' developed as prototypes for community modules that use dynamic web pages to present data obtained on-demand from third-party servers via RESTful web services. Designed for sustainability, the Arabidopsis Information Portal strategy exploits existing scientific computing infrastructure, adopts a practical mixture of data integration technologies and encourages collaborative enhancement of the resource by its user community.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

