Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells
- PMID: 25414332
- PMCID: PMC4330342
- DOI: 10.1093/nar/gku1246
Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells
Abstract
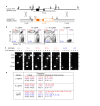
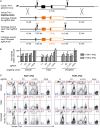
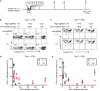
Sequence-specific nucleases such as TALEN and the CRISPR/Cas9 system have so far been used to disrupt, correct or insert transgenes at precise locations in mammalian genomes. We demonstrate efficient 'knock-in' targeted replacement of multi-kilobase genes in human induced pluripotent stem cells (iPSC). Using a model system replacing endogenous human genes with their mouse counterpart, we performed a comprehensive study of targeting vector design parameters for homologous recombination. A 2.7 kilobase (kb) homozygous gene replacement was achieved in up to 11% of iPSC without selection. The optimal homology arm length was around 2 kb, with homology length being especially critical on the arm not adjacent to the cut site. Homologous sequence inside the cut sites was detrimental to targeting efficiency, consistent with a synthesis-dependent strand annealing (SDSA) mechanism. Using two nuclease sites, we observed a high degree of gene excisions and inversions, which sometimes occurred more frequently than indel mutations. While homozygous deletions of 86 kb were achieved with up to 8% frequency, deletion frequencies were not solely a function of nuclease activity and deletion size. Our results analyzing the optimal parameters for targeting vector design will inform future gene targeting efforts involving multi-kilobase gene segments, particularly in human iPSC.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
References
-
- Bollag R.J., Waldman A.S., Liskay R.M. Homologous recombination in mammalian cells. Annu. Rev. Genet. 1989;23:199–225. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

