ProteomeScout: a repository and analysis resource for post-translational modifications and proteins
- PMID: 25414335
- PMCID: PMC4383955
- DOI: 10.1093/nar/gku1154
ProteomeScout: a repository and analysis resource for post-translational modifications and proteins
Abstract
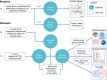
ProteomeScout (https://proteomescout.wustl.edu) is a resource for the study of proteins and their post-translational modifications (PTMs) consisting of a database of PTMs, a repository for experimental data, an analysis suite for PTM experiments, and a tool for visualizing the relationships between complex protein annotations. The PTM database is a compendium of public PTM data, coupled with user-uploaded experimental data. ProteomeScout provides analysis tools for experimental datasets, including summary views and subset selection, which can identify relationships within subsets of data by testing for statistically significant enrichment of protein annotations. Protein annotations are incorporated in the ProteomeScout database from external resources and include terms such as Gene Ontology annotations, domains, secondary structure and non-synonymous polymorphisms. These annotations are available in the database download, in the analysis tools and in the protein viewer. The protein viewer allows for the simultaneous visualization of annotations in an interactive web graphic, which can be exported in Scalable Vector Graphics (SVG) format. Finally, quantitative data measurements associated with public experiments are also easily viewable within protein records, allowing researchers to see how PTMs change across different contexts. ProteomeScout should prove useful for protein researchers and should benefit the proteomics community by providing a stable repository for PTM experiments.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
-
- Witze E.S., Old W.M., Resing K.A., Ahn N.G. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods. 2007;4:798–806. - PubMed
-
- Hornbeck P.V., Kornhauser J.M., Tkachev S., Zhang B., Skrzypek E., Murray B., Latham V., Sullivan M. PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012;40:D261–D270. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

