In silico Derivation of HLA-Specific Alloreactivity Potential from Whole Exome Sequencing of Stem-Cell Transplant Donors and Recipients: Understanding the Quantitative Immunobiology of Allogeneic Transplantation
- PMID: 25414699
- PMCID: PMC4222229
- DOI: 10.3389/fimmu.2014.00529
In silico Derivation of HLA-Specific Alloreactivity Potential from Whole Exome Sequencing of Stem-Cell Transplant Donors and Recipients: Understanding the Quantitative Immunobiology of Allogeneic Transplantation
Abstract
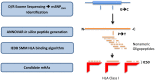
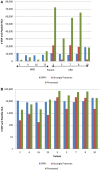
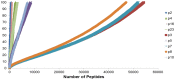
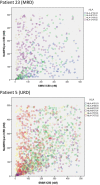
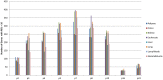
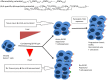
Donor T-cell mediated graft versus host (GVH) effects may result from the aggregate alloreactivity to minor histocompatibility antigens (mHA) presented by the human leukocyte antigen (HLA) molecules in each donor-recipient pair undergoing stem-cell transplantation (SCT). Whole exome sequencing has previously demonstrated a large number of non-synonymous single nucleotide polymorphisms (SNP) present in HLA-matched recipients of SCT donors (GVH direction). The nucleotide sequence flanking each of these SNPs was obtained and the amino acid sequence determined. All the possible nonameric peptides incorporating the variant amino acid resulting from these SNPs were interrogated in silico for their likelihood to be presented by the HLA class I molecules using the Immune Epitope Database stabilized matrix method (SMM) and NetMHCpan algorithms. The SMM algorithm predicted that a median of 18,396 peptides weakly bound HLA class I molecules in individual SCT recipients, and 2,254 peptides displayed strong binding. A similar library of presented peptides was identified when the data were interrogated using the NetMHCpan algorithm. The bioinformatic algorithm presented here demonstrates that there may be a high level of mHA variation in HLA-matched individuals, constituting a HLA-specific alloreactivity potential.
Keywords: HLA; alloreactivity potential; minor histocompatibility antigen; stem-cell transplant; whole exome sequencing.
Figures

References
-
- Toor AA, Sabo RT, Chung HM, Roberts C, Manjili RH, Song S, et al. Favorable outcomes in patients with high donor-derived T cell count after in vivo T cell-depleted reduced-intensity allogeneic stem cell transplantation. Biol Blood Marrow Transplant (2012) 18(5):794–794.10.1016/j.bbmt.2011.10.011 - DOI - PMC - PubMed
-
- Weisdorf DJ, Nelson G, Lee SJ, Haagenson M, Spellman S, Antin JH, et al. Sibling versus unrelated donor allogeneic hematopoietic cell transplantation for chronic myelogenous leukemia: refined HLA matching reveals more graft-versus-host disease but not less relapse. Biol Blood Marrow Transplant (2009) 15(11):1475–8.10.1016/j.bbmt.2009.06.016 - DOI - PMC - PubMed
-
- Valcarcel D, Sierra J, Wang T, Kan F, Gupta V, Hale GA, et al. One-antigen mismatched related versus HLA-matched unrelated donor hematopoietic stem cell transplantation in adults with acute leukemia: center for international blood and marrow transplant research results in the era of molecular HLA typing. Biol Blood Marrow Transplant (2011) 17(5):640–8.10.1016/j.bbmt.2010.07.022 - DOI - PMC - PubMed
-
- Brunstein CG, Fuchs EJ, Carter SL, Karanes C, Costa LJ, Wu J, et al. Alternative donor transplantation after reduced intensity conditioning: results of parallel phase 2 trials using partially HLA-mismatched related bone marrow or unrelated double umbilical cord blood grafts. Blood (2011) 118(2):282–8.10.1182/blood-2011-03-344853 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

