The krebs cycle enzyme α-ketoglutarate decarboxylase is an essential glycosomal protein in bloodstream African trypanosomes
- PMID: 25416237
- PMCID: PMC4346564
- DOI: 10.1128/EC.00214-14
The krebs cycle enzyme α-ketoglutarate decarboxylase is an essential glycosomal protein in bloodstream African trypanosomes
Abstract
α-Ketoglutarate decarboxylase (α-KDE1) is a Krebs cycle enzyme found in the mitochondrion of the procyclic form (PF) of Trypanosoma brucei. The bloodstream form (BF) of T. brucei lacks a functional Krebs cycle and relies exclusively on glycolysis for ATP production. Despite the lack of a functional Krebs cycle, α-KDE1 was expressed in BF T. brucei and RNA interference knockdown of α-KDE1 mRNA resulted in rapid growth arrest and killing. Cell death was preceded by progressive swelling of the flagellar pocket as a consequence of recruitment of both flagellar and plasma membranes into the pocket. BF T. brucei expressing an epitope-tagged copy of α-KDE1 showed localization to glycosomes and not the mitochondrion. We used a cell line transfected with a reporter construct containing the N-terminal sequence of α-KDE1 fused to green fluorescent protein to examine the requirements for glycosome targeting. We found that the N-terminal 18 amino acids of α-KDE1 contain overlapping mitochondrion- and peroxisome-targeting sequences and are sufficient to direct localization to the glycosome in BF T. brucei. These results suggest that α-KDE1 has a novel moonlighting function outside the mitochondrion in BF T. brucei.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures

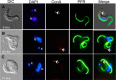
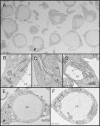

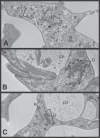

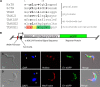
Similar articles
-
Identification and characterization of three peroxins--PEX6, PEX10 and PEX12--involved in glycosome biogenesis in Trypanosoma brucei.Biochim Biophys Acta. 2006 Jan;1763(1):6-17. doi: 10.1016/j.bbamcr.2005.11.002. Epub 2005 Dec 7. Biochim Biophys Acta. 2006. PMID: 16388862
-
Characterization of the role of the receptors PEX5 and PEX7 in the import of proteins into glycosomes of Trypanosoma brucei.Biochim Biophys Acta. 2007 Apr;1773(4):521-35. doi: 10.1016/j.bbamcr.2007.01.006. Epub 2007 Jan 20. Biochim Biophys Acta. 2007. PMID: 17320990
-
An internal sequence targets Trypanosoma brucei triosephosphate isomerase to glycosomes.Mol Biochem Parasitol. 2010 May;171(1):45-9. doi: 10.1016/j.molbiopara.2010.01.002. Epub 2010 Feb 4. Mol Biochem Parasitol. 2010. PMID: 20138091
-
Targeting proteins to the glycosomes of African trypanosomes.Annu Rev Microbiol. 1994;48:105-38. doi: 10.1146/annurev.mi.48.100194.000541. Annu Rev Microbiol. 1994. PMID: 7826002 Review.
-
In or out? On the tightness of glycosomal compartmentalization of metabolites and enzymes in Trypanosoma brucei.Mol Biochem Parasitol. 2014 Nov;198(1):18-28. doi: 10.1016/j.molbiopara.2014.11.004. Epub 2014 Dec 2. Mol Biochem Parasitol. 2014. PMID: 25476771 Review.
Cited by
-
Physiological and proteomic profiles of Trypanosoma brucei rhodesiense parasite isolated from suramin responsive and non-responsive HAT patients in Busoga, Uganda.Int J Parasitol Drugs Drug Resist. 2021 Apr;15:57-67. doi: 10.1016/j.ijpddr.2021.02.001. Epub 2021 Feb 9. Int J Parasitol Drugs Drug Resist. 2021. PMID: 33588295 Free PMC article.
-
Gluconeogenesis using glycerol as a substrate in bloodstream-form Trypanosoma brucei.PLoS Pathog. 2018 Dec 27;14(12):e1007475. doi: 10.1371/journal.ppat.1007475. eCollection 2018 Dec. PLoS Pathog. 2018. PMID: 30589893 Free PMC article.
-
Isocitrate Lyase and Succinate Semialdehyde Dehydrogenase Mediate the Synthesis of α-Ketoglutarate in Pseudomonas fluorescens.Front Microbiol. 2019 Aug 23;10:1929. doi: 10.3389/fmicb.2019.01929. eCollection 2019. Front Microbiol. 2019. PMID: 31507554 Free PMC article.
-
Extracellular Vesicles from Trypanosoma brucei Mediate Virulence Factor Transfer and Cause Host Anemia.Cell. 2016 Jan 14;164(1-2):246-257. doi: 10.1016/j.cell.2015.11.051. Cell. 2016. PMID: 26771494 Free PMC article.
-
Ethyl Pyruvate Emerges as a Safe and Fast Acting Agent against Trypanosoma brucei by Targeting Pyruvate Kinase Activity.PLoS One. 2015 Sep 4;10(9):e0137353. doi: 10.1371/journal.pone.0137353. eCollection 2015. PLoS One. 2015. PMID: 26340747 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

