Molecular characterization of H3N2 influenza A viruses isolated from Ontario swine in 2011 and 2012
- PMID: 25416300
- PMCID: PMC4245826
- DOI: 10.1186/s12985-014-0194-z
Molecular characterization of H3N2 influenza A viruses isolated from Ontario swine in 2011 and 2012
Abstract
Background: Data about molecular diversity of commonly circulating type A influenza viruses in Ontario swine are scarce. Yet, this information is essential for surveillance of animal and public health, vaccine updates, and for understanding virus evolution and its large-scale spread.
Methods: The study population consisted of 21 swine herds with clinical problems due to respiratory disease. Nasal swabs from individual pigs were collected and tested by virus isolation in MDCK cells and by rtRT-PCR. All eight segments of 10 H3N2 viruses were sequenced using high-throughput sequencing and molecularly characterized.
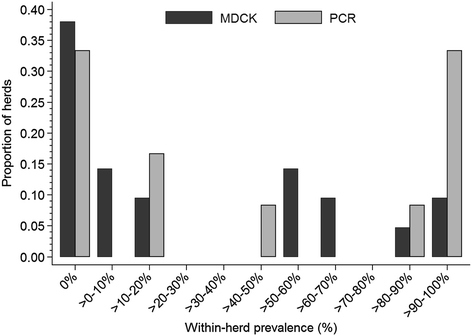
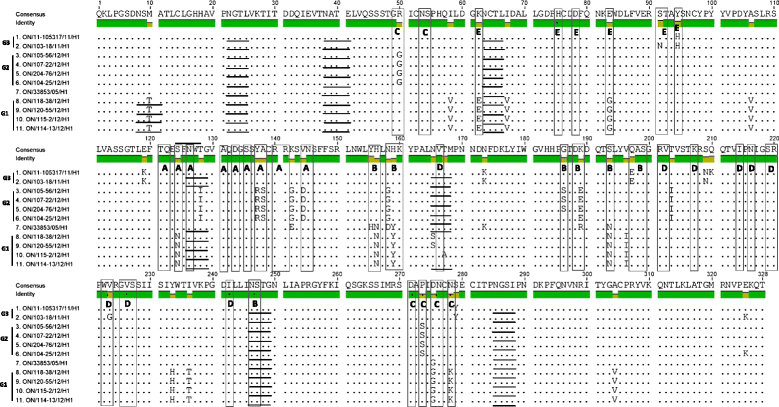
Results: Within-herd prevalence ranged between 2 and 100%. Structurally, Ontario H3N2 viruses could be classified into three different groups. Group 1 was the most similar to the original trH3N2 virus from 2005. Group 2 was the most similar to the Ontario turkey H3N2 isolates with PB1 and NS genes originating from trH3N2 virus and M, PB2, PA and NP genes originating from the A(H1N1)pdm09 virus. All Group 3 internal genes were genetically related to A(H1N1)pdm09. Analysis of antigenic sites of HA1 showed that Group 1 had 8 aa changes within 4 antigenic sites, A(1), B(3), C(2) and E(2). The Group 2 viruses had 8 aa changes within 3 antigenic sites A(3), B(3) and C(2), while Group 3 viruses had 4 aa changes within 3 antigenic sites, B(1), D(1) and E(2), when compared to the cluster IV H3N2 virus [A/swine/Ontario/33853/2005/(H3N2)].
Conclusions: The characterization of the Ontario H3N2 viruses clearly indicates reassortment of gene segments between the North American swine trH3N2 from cluster IV and the A(H1N1)pdm09 virus.
Figures



References
-
- Shaw ML, Palese P. Orthomyxoviruses. In: Knipe DM, Howley PM, editors. Field Virology. 6. Philahelphia, PA, USA: Lippincott Williams & Wilkins; 2014. pp. 1151–1185.
-
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA, Donis RO: New world bats harbor diverse influenza A viruses.PLoS Pathog 2013, 9(10):e1003657. - PMC - PubMed
-
- Anonymous: Canadian Hog Farms. [http://www.cpc-ccp.com/canadian_hog_farms.php] Accessed 2014/09/06
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

