Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle
- PMID: 25416771
- PMCID: PMC4292475
- DOI: 10.1128/AEM.03018-14
Few highly abundant operational taxonomic units dominate within rumen methanogenic archaeal species in New Zealand sheep and cattle
Abstract
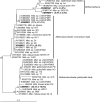
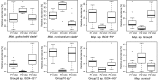
Sequencing and analyses of 16S rRNA gene amplicons were performed to estimate the composition of the rumen methanogen community in 252 samples from eight cohorts of sheep and cattle, separated into 16 different sample groups by diet, and to determine which methanogens are most prominent in the rumens of farmed New Zealand ruminants. Methanobacteriales (relative abundance ± standard deviation, 89.6% ± 9.8%) and Methanomassiliicoccales (10.4% ± 9.8%) were the two major orders and contributed 99.98% (±0.1%) to the rumen methanogen communities in the samples. Sequences from Methanobacteriales were almost entirely from only four different species (or clades of very closely related species). Each was detectable in at least 89% of the samples. These four species or clades were the Methanobrevibacter gottschalkii clade and Methanobrevibacter ruminantium clade with a mean abundance of 42.4% (±19.5% standard deviation) and 32.9% (±18.8%), respectively, and Methanosphaera sp. ISO3-F5 (8.2% ± 6.7%) and Methanosphaera sp. group5 (5.6% ± 5.7%). These four species or clades appeared to be primarily represented by only one or, in one case, two dominant sequence types per species or clade when the sequences were grouped into operational taxonomic units (OTUs) at 99% sequence identity. The mean relative abundance of Methanomassiliicoccales in the samples was relatively low but exceeded 40% in some of the treatment groups. Animal feed affected the apparent methanogen community structure of both orders, as evident from differences in relative abundances of the major OTUs in animals under different feeding regimens.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures



References
-
- Yusuf RO, Noor ZZ, Abba AH, Hassan MAA, Din MFM. 2012. Methane emission by sectors: a comprehensive review of emission sources and mitigation methods. Renew Sustain Energy Rev 16:5059–5070. doi:10.1016/j.rser.2012.04.008. - DOI
-
- Clark H, Kelliher F, Pinares-Patino C. 2011. Reducing CH4 emissions from grazing ruminants in New Zealand: challenges and opportunities. Asian Australas J Anim Sci 24:295–302. doi:10.5713/ajas.2011.r.04. - DOI
-
- Joblin KN. 2005. Methanogenic archaea, p 47–53. In Makkar HPS, McSweeney CS (ed), Methods in gut microbial ecology for ruminants. Springer, Dordrecht, The Netherlands.
-
- Iino T, Tamaki H, Tamazawa S, Ueno Y, Ohkuma M, Suzuki K, Igarashi Y, Haruta S. 2013. Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microbes Environ 28:244–250. doi:10.1264/jsme2.ME12189. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

