Structural model of weak binding actomyosin in the prepowerstroke state
- PMID: 25416786
- PMCID: PMC4340411
- DOI: 10.1074/jbc.M114.606665
Structural model of weak binding actomyosin in the prepowerstroke state
Abstract
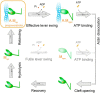
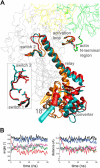
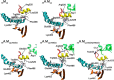
We present the first in silico model of the weak binding actomyosin in the initial powerstroke state, representing the actin binding-induced major structural changes in myosin. First, we docked an actin trimer to prepowerstroke myosin then relaxed the complex by a 100-ns long unrestrained molecular dynamics. In the first few nanoseconds, actin binding induced an extra primed myosin state, i.e. the further priming of the myosin lever by 18° coupled to a further closure of switch 2 loop. We demonstrated that actin induces the extra primed state of myosin specifically through the actin N terminus-activation loop interaction. The applied in silico methodology was validated by forming rigor structures that perfectly fitted into an experimentally determined EM map of the rigor actomyosin. Our results unveiled the role of actin in the powerstroke by presenting that actin moves the myosin lever to the extra primed state that leads to the effective lever swing.
Keywords: Actin; Enzyme Cycle; Enzyme Kinetics; Enzyme Mechanism; Molecular Dynamics; Myosin; Powerstroke; Protein Structure; Structural Model.
© 2015 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



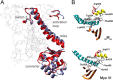
References
-
- Pollard T. D. (2000) Reflections on a quarter century of research on contractile systems. Trends Biochem. Sci. 25, 607–611 - PubMed
-
- Geeves M. A., Holmes K. C. (2005) The molecular mechanism of muscle contraction. Adv. Protein Chem. 71, 161–193 - PubMed
-
- Sweeney H. L., Houdusse A. (2010) Structural and functional insights into the myosin motor mechanism. Annu. Rev. Biophys. 39, 539–557 - PubMed
-
- Gyimesi M., Kintses B., Bodor A., Perczel A., Fischer S., Bagshaw C. R., Málnási-Csizmadia A. (2008) The mechanism of the reverse recovery step, phosphate release, and actin activation of Dictyostelium myosin II. J. Biol. Chem. 283, 8153–8163 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

