Unraveling the biology of a fungal meningitis pathogen using chemical genetics
- PMID: 25416953
- PMCID: PMC4243055
- DOI: 10.1016/j.cell.2014.10.044
Unraveling the biology of a fungal meningitis pathogen using chemical genetics
Abstract
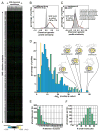
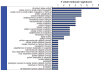
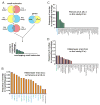
The fungal meningitis pathogen Cryptococcus neoformans is a central driver of mortality in HIV/AIDS. We report a genome-scale chemical genetic data map for this pathogen that quantifies the impact of 439 small-molecule challenges on 1,448 gene knockouts. We identified chemical phenotypes for 83% of mutants screened and at least one genetic response for each compound. C. neoformans chemical-genetic responses are largely distinct from orthologous published profiles of Saccharomyces cerevisiae, demonstrating the importance of pathogen-centered studies. We used the chemical-genetic matrix to predict novel pathogenicity genes, infer compound mode of action, and to develop an algorithm, O2M, that predicts antifungal synergies. These predictions were experimentally validated, thereby identifying virulence genes, a molecule that triggers G2/M arrest and inhibits the Cdc25 phosphatase, and many compounds that synergize with the antifungal drug fluconazole. Our work establishes a chemical-genetic foundation for approaching an infection responsible for greater than one-third of AIDS-related deaths.
Figures



References
-
- Anderson JB. Evolution of antifungal-drug resistance: mechanisms and pathogen fitness. Nature Reviews Microbiology. 2005;3:547–556. - PubMed
-
- Armstrong-James D, Meintjes G, Brown GD. A neglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol. 2014;22:120–127. - PubMed
-
- Broek D, Bartlett R, Crawford K, Nurse P. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature. 1991;349:388–393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

