An integrated approach reveals regulatory controls on bacterial translation elongation
- PMID: 25416955
- PMCID: PMC4243059
- DOI: 10.1016/j.cell.2014.10.043
An integrated approach reveals regulatory controls on bacterial translation elongation
Abstract
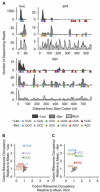
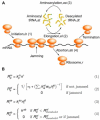
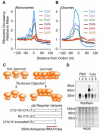
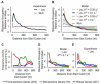
Ribosomes elongate at a nonuniform rate during translation. Theoretical models and experiments disagree on the in vivo determinants of elongation rate and the mechanism by which elongation rate affects protein levels. To resolve this conflict, we measured transcriptome-wide ribosome occupancy under multiple conditions and used it to formulate a whole-cell model of translation in E. coli. Our model predicts that elongation rates at most codons during nutrient-rich growth are not limited by the intracellular concentrations of aminoacyl-tRNAs. However, elongation pausing during starvation for single amino acids is highly sensitive to the kinetics of tRNA aminoacylation. We further show that translation abortion upon pausing accounts for the observed ribosome occupancy along mRNAs during starvation. Abortion reduces global protein synthesis, but it enhances the translation of a subset of mRNAs. These results suggest a regulatory role for aminoacylation and abortion during stress, and our study provides an experimentally constrained framework for modeling translation.
Figures


References
-
- Abo T, Ueda K, Sunohara T, Ogawa K, Aiba H. SsrA-mediated protein tagging in the presence of miscoding drugs and its physiological role in Escherichia coli. Genes Cells. 2002;7:629–638. - PubMed
-
- Bilgin N, Ehrenberg M. Mutations in 23 S Ribosomal RNA Perturb Transfer RNA Selection and can Lead to Streptomycin Dependence. J. Mol. Biol. 1994;235:813–824. - PubMed
-
- Bordbar A, Monk J, King Z, Palsson B. Constraint-based models predict metabolic and associated cellular functions. Nat Rev Genet. 2014;15 - PubMed
-
- Bremer H, Dennis P. Modulation of Chemical Composition and Other Parameters of the Cell at Different Exponential Growth Rates. EcoSal Plus. 2008 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

