Deterministic progenitor behavior and unitary production of neurons in the neocortex
- PMID: 25417155
- PMCID: PMC4225456
- DOI: 10.1016/j.cell.2014.10.027
Deterministic progenitor behavior and unitary production of neurons in the neocortex
Abstract
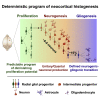
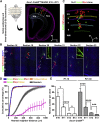
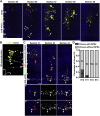
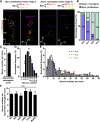
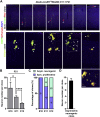
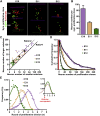
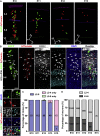
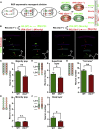
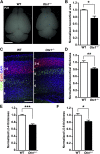
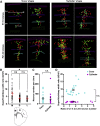
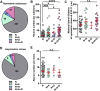
Radial glial progenitors (RGPs) are responsible for producing nearly all neocortical neurons. To gain insight into the patterns of RGP division and neuron production, we quantitatively analyzed excitatory neuron genesis in the mouse neocortex using Mosaic Analysis with Double Markers, which provides single-cell resolution of progenitor division patterns and potential in vivo. We found that RGPs progress through a coherent program in which their proliferative potential diminishes in a predictable manner. Upon entry into the neurogenic phase, individual RGPs produce ?8-9 neurons distributed in both deep and superficial layers, indicating a unitary output in neuronal production. Removal of OTX1, a transcription factor transiently expressed in RGPs, results in both deep- and superficial-layer neuron loss and a reduction in neuronal unit size. Moreover, ?1/6 of neurogenic RGPs proceed to produce glia. These results suggest that progenitor behavior and histogenesis in the mammalian neocortex conform to a remarkably orderly and deterministic program.
Figures




References
-
- Acampora D., Mazan S., Avantaggiato V., Barone P., Tuorto F., Lallemand Y., Brûlet P., Simeone A. Epilepsy and brain abnormalities in mice lacking the Otx1 gene. Nat. Genet. 1996;14:218–222. - PubMed
-
- Angevine J.B., Jr., Sidman R.L. Autoradiographic study of cell migration during histogenesis of cerebral cortex in the mouse. Nature. 1961;192:766–768. - PubMed
-
- Anthony T.E., Klein C., Fishell G., Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. - PubMed
-
- Betizeau M., Cortay V., Patti D., Pfister S., Gautier E., Bellemin-Ménard A., Afanassieff M., Huissoud C., Douglas R.J., Kennedy H., Dehay C. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron. 2013;80:442–457. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

