Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation
- PMID: 25417163
- PMCID: PMC4243038
- DOI: 10.1016/j.cell.2014.09.055
Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation
Abstract
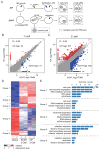
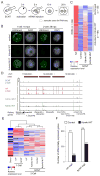
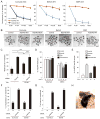
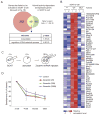
Mammalian oocytes can reprogram somatic cells into a totipotent state enabling animal cloning through somatic cell nuclear transfer (SCNT). However, the majority of SCNT embryos fail to develop to term due to undefined reprogramming defects. Here, we identify histone H3 lysine 9 trimethylation (H3K9me3) of donor cell genome as a major barrier for efficient reprogramming by SCNT. Comparative transcriptome analysis identified reprogramming resistant regions (RRRs) that are expressed normally at 2-cell mouse embryos generated by in vitro fertilization (IVF) but not SCNT. RRRs are enriched for H3K9me3 in donor somatic cells and its removal by ectopically expressed H3K9me3 demethylase Kdm4d not only reactivates the majority of RRRs, but also greatly improves SCNT efficiency. Furthermore, use of donor somatic nuclei depleted of H3K9 methyltransferases markedly improves SCNT efficiency. Our study thus identifies H3K9me3 as a critical epigenetic barrier in SCNT-mediated reprogramming and provides a promising approach for improving mammalian cloning efficiency.
Figures


References
-
- Akagi S, Matsukawa K, Mizutani E, Fukunari K, Kaneda M, Watanabe S, Takahashi S. Treatment with a histone deacetylase inhibitor after nuclear transfer improves the preimplantation development of cloned bovine embryos. J Reprod Dev. 2011;57:120–126. - PubMed
-
- Bannister aJ, Zegerman P, Partridge JF, Miska Ea, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

