Toehold switches: de-novo-designed regulators of gene expression
- PMID: 25417166
- PMCID: PMC4265554
- DOI: 10.1016/j.cell.2014.10.002
Toehold switches: de-novo-designed regulators of gene expression
Abstract
Efforts to construct synthetic networks in living cells have been hindered by the limited number of regulatory components that provide wide dynamic range and low crosstalk. Here, we report a class of de-novo-designed prokaryotic riboregulators called toehold switches that activate gene expression in response to cognate RNAs with arbitrary sequences. Toehold switches provide a high level of orthogonality and can be forward engineered to provide average dynamic range above 400. We show that switches can be integrated into the genome to regulate endogenous genes and use them as sensors that respond to endogenous RNAs. We exploit the orthogonality of toehold switches to regulate 12 genes independently and to construct a genetic circuit that evaluates 4-input AND logic. Toehold switches, with their wide dynamic range, orthogonality, and programmability, represent a versatile and powerful platform for regulation of translation, offering diverse applications in molecular biology, synthetic biology, and biotechnology.
Figures




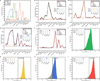
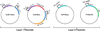

Comment in
-
Synthetic biology: Toehold gene switches make big footprints.Nature. 2014 Dec 18;516(7531):333-4. doi: 10.1038/516333a. Nature. 2014. PMID: 25519125 No abstract available.
References
-
- Auslaender S, Auslaender D, Mueller M, Wieland M, Fussenegger M. Programmable single-cell mammalian biocomputers. Nature. 2012;487:123–127. - PubMed
-
- Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. - PubMed
-
- Brantl S, Wagner EGH. Antisense RNA-mediated transcriptional attenuation: an in vitro study of plasmid pT181. Mol. Microbiol. 2000;35:1469–1482. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

