Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient
- PMID: 25418686
- PMCID: PMC4223161
- DOI: 10.1186/s13059-014-0490-3
Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient
Abstract
Background: Mycobacterium tuberculosis is characterized by a low mutation rate and a lack of genetic recombination. Yet, the rise of extensively resistant strains paints a picture of a microbe with an impressive adaptive potential. Here we describe the first documented case of extensively drug-resistant tuberculosis evolved from a susceptible ancestor within a single patient.
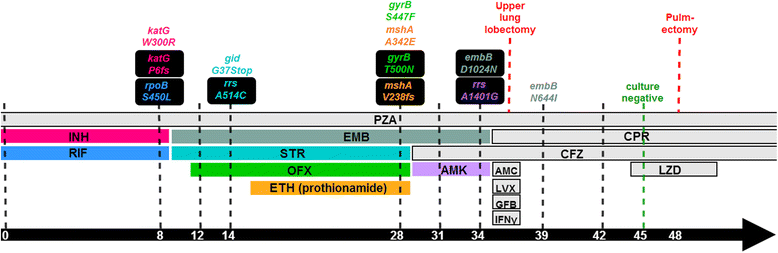
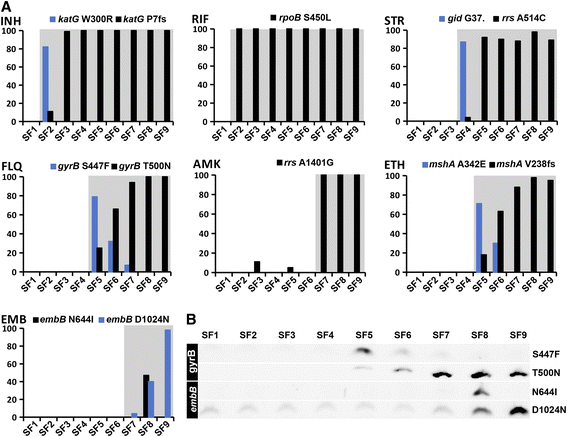
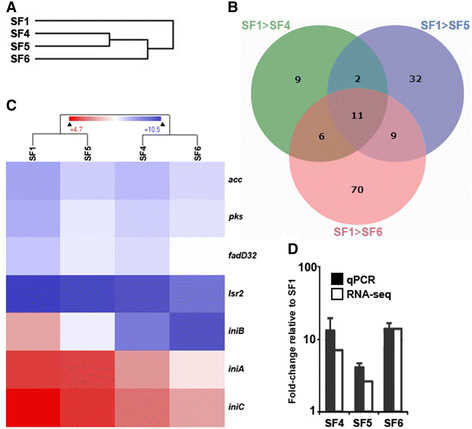
Results: Genome sequences of nine serial M. tuberculosis isolates from the same patient uncovered a dramatic turnover of competing lineages driven by the emergence, and subsequent fixation or loss of single nucleotide polymorphisms. For most drugs, resistance arose through independent emergence of mutations in more than one clone, of which only one ultimately prevailed as the clone carrying it expanded, displacing the other clones in the process. The vast majority of mutations identified over 3.5 years were either involved in drug resistance or hitchhiking in the genetic background of these. Additionally, RNA-sequencing of isolates grown in the absence of drug challenge revealed that the efflux-associated iniBAC operon was up-regulated over time, whereas down-regulated genes include those involved in mycolic acid synthesis.
Conclusions: We observed both rapid acquisitions of resistance to antimicrobial compounds mediated by individual mutations as well as a gradual increase in fitness in the presence of antibiotics, likely driven by stable gene expression reprogramming. The rapid turnover of resistance mutations and hitchhiking neutral mutations has major implications for inferring tuberculosis transmission events in situations where drug resistance evolves within transmission chains.
Figures


References
-
- Ford CB, Shah RR, Maeda MK, Gagneux S, Murray MB, Cohen T, Johnston JC, Gardy J, Lipsitch M, Fortune SM. Mycobacterium tuberculosis mutation rate estimates from different lineages predict substantial differences in the emergence of drug-resistant tuberculosis. Nat Genet. 2013;45:784–790. doi: 10.1038/ng.2656. - DOI - PMC - PubMed
-
- Klopper M, Warren RM, Hayes C, van Pittius NCG, Streicher EM, Müller B, Sirgel FA, Chabula-Nxiweni M, Hoosain E, Coetzee G, David van Helden P, Victor TC, Trollip AP. Emergence and spread of extensively and totally drug-resistant tuberculosis, South Africa. Emerg Infect Dis. 2013;19:449–455. doi: 10.3201/eid1903.120246. - DOI - PMC - PubMed
-
- WHO: Weekly Epidemiological Record No 45. Volume 81. 2006, 425–432 [http://www.who.int/wer/2006/wer8145/en/]
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

