A community computational challenge to predict the activity of pairs of compounds
- PMID: 25419740
- PMCID: PMC4399794
- DOI: 10.1038/nbt.3052
A community computational challenge to predict the activity of pairs of compounds
Abstract
Recent therapeutic successes have renewed interest in drug combinations, but experimental screening approaches are costly and often identify only small numbers of synergistic combinations. The DREAM consortium launched an open challenge to foster the development of in silico methods to computationally rank 91 compound pairs, from the most synergistic to the most antagonistic, based on gene-expression profiles of human B cells treated with individual compounds at multiple time points and concentrations. Using scoring metrics based on experimental dose-response curves, we assessed 32 methods (31 community-generated approaches and SynGen), four of which performed significantly better than random guessing. We highlight similarities between the methods. Although the accuracy of predictions was not optimal, we find that computational prediction of compound-pair activity is possible, and that community challenges can be useful to advance the field of in silico compound-synergy prediction.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

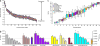


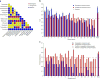
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- 5U54CA121852-08/CA/NCI NIH HHS/United States
- U01 CA168426/CA/NCI NIH HHS/United States
- 104104/Wellcome Trust/United Kingdom
- 5R01CA152301/CA/NCI NIH HHS/United States
- U01 HL111566/HL/NHLBI NIH HHS/United States
- R01 CA152301/CA/NCI NIH HHS/United States
- P50 CA058207/CA/NCI NIH HHS/United States
- U54 CA121852/CA/NCI NIH HHS/United States
- 3U01HL111566-02/HL/NHLBI NIH HHS/United States
- U54 CA112970/CA/NCI NIH HHS/United States
- R01 GM071966/GM/NIGMS NIH HHS/United States
- U01 CA164184/CA/NCI NIH HHS/United States
- Howard Hughes Medical Institute/United States
- R01 GM081871/GM/NIGMS NIH HHS/United States
- 1U01CA164184-02/CA/NCI NIH HHS/United States
- T32 HG003284/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

