Selective conversion of fibroblasts into peripheral sensory neurons
- PMID: 25420069
- PMCID: PMC4466122
- DOI: 10.1038/nn.3887
Selective conversion of fibroblasts into peripheral sensory neurons
Abstract
Humans and mice detect pain, itch, temperature, pressure, stretch and limb position via signaling from peripheral sensory neurons. These neurons are divided into three functional classes (nociceptors/pruritoceptors, mechanoreceptors and proprioceptors) that are distinguished by their selective expression of TrkA, TrkB or TrkC receptors, respectively. We found that transiently coexpressing Brn3a with either Ngn1 or Ngn2 selectively reprogrammed human and mouse fibroblasts to acquire key properties of these three classes of sensory neurons. These induced sensory neurons (iSNs) were electrically active, exhibited distinct sensory neuron morphologies and matched the characteristic gene expression patterns of endogenous sensory neurons, including selective expression of Trk receptors. In addition, we found that calcium-imaging assays could identify subsets of iSNs that selectively responded to diverse ligands known to activate itch- and pain-sensing neurons. These results offer a simple and rapid means for producing genetically diverse human sensory neurons suitable for drug screening and mechanistic studies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

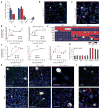


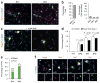
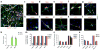

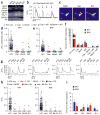
References
-
- Finnerup NB, Sindrup SH, Jensen TS. The evidence for pharmacological treatment of neuropathic pain. Pain. 2010;150:573–581. - PubMed
-
- Scholz J, Woolf CJ. Can we conquer pain? Nat Neurosci. 2002;5:1062–1067. - PubMed
-
- Koeppen AH, et al. The dorsal root ganglion in Friedreich’s ataxia. Acta Neuropathol. 2009;118:763–776. - PubMed
-
- Riera CE, et al. TRPV1 pain receptors regulate longevity and metabolism by neuropeptide signaling. Cell. 2014;157:1023–1036. - PubMed
-
- Dib-Hajj SD, Yang Y, Black JA, Waxman SG. The Na(V)1.7 sodium channel: from molecule to man. Nat Rev Neurosci. 2013;14:49–62. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

