Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley
- PMID: 25420105
- PMCID: PMC4242610
- DOI: 10.1371/journal.pone.0113120
Genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley
Erratum in
-
Correction: genetic dissection of photoperiod response based on GWAS of pre-anthesis phase duration in spring barley.PLoS One. 2015 Apr 7;10(4):e0123748. doi: 10.1371/journal.pone.0123748. eCollection 2015. PLoS One. 2015. PMID: 25849431 Free PMC article. No abstract available.
Abstract
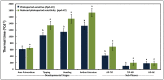
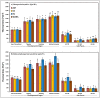
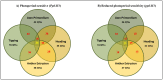
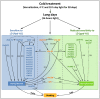
Heading time is a complex trait, and natural variation in photoperiod responses is a major factor controlling time to heading, adaptation and grain yield. In barley, previous heading time studies have been mainly conducted under field conditions to measure total days to heading. We followed a novel approach and studied the natural variation of time to heading in a world-wide spring barley collection (218 accessions), comprising of 95 photoperiod-sensitive (Ppd-H1) and 123 accessions with reduced photoperiod sensitivity (ppd-H1) to long-day (LD) through dissecting pre-anthesis development into four major stages and sub-phases. The study was conducted under greenhouse (GH) conditions (LD; 16/8 h; ∼20/∼16°C day/night). Genotyping was performed using a genome-wide high density 9K single nucleotide polymorphisms (SNPs) chip which assayed 7842 SNPs. We used the barley physical map to identify candidate genes underlying genome-wide association scans (GWAS). GWAS for pre-anthesis stages/sub-phases in each photoperiod group provided great power for partitioning genetic effects on floral initiation and heading time. In addition to major genes known to regulate heading time under field conditions, several novel QTL with medium to high effects, including new QTL having major effects on developmental stages/sub-phases were found to be associated in this study. For example, highly associated SNPs tagged the physical regions around HvCO1 (barley CONSTANS1) and BFL (BARLEY FLORICAULA/LEAFY) genes. Based upon our GWAS analysis, we propose a new genetic network model for each photoperiod group, which includes several newly identified genes, such as several HvCO-like genes, belonging to different heading time pathways in barley.
Conflict of interest statement
Figures





References
-
- Andres F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nature Reviews Genetics 13:627–639. - PubMed
-
- Purugganan MD, Fuller DQ (2009) The nature of selection during plant domestication. Nature 457:843–848. - PubMed
-
- Alqudah AM, Schnurbusch T (2014) Awn primordium to tipping is the most decisive developmental phase for spikelet survival in barley. Functional Plant Biology 41:424–436. - PubMed
-
- Sreenivasulu N, Schnurbusch T (2012) A genetic playground for enhancing grain number in cereals. Trends in Plant Science 17:91–101. - PubMed
-
- Borràs G, Romagosa I, van Eeuwijk F, Slafer GA (2009) Genetic variability in duration of pre-heading phases and relationships with leaf appearance and tillering dynamics in a barley population. Field Crops Research 113:95–104.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

