Synergy: a web resource for exploring gene regulation in Synechocystis sp. PCC6803
- PMID: 25420108
- PMCID: PMC4242644
- DOI: 10.1371/journal.pone.0113496
Synergy: a web resource for exploring gene regulation in Synechocystis sp. PCC6803
Abstract
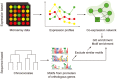
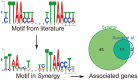
Despite being a highly studied model organism, most genes of the cyanobacterium Synechocystis sp. PCC 6803 encode proteins with completely unknown function. To facilitate studies of gene regulation in Synechocystis, we have developed Synergy (http://synergy.plantgenie.org), a web application integrating co-expression networks and regulatory motif analysis. Co-expression networks were inferred from publicly available microarray experiments, while regulatory motifs were identified using a phylogenetic footprinting approach. Automatically discovered motifs were shown to be enriched in the network neighborhoods of regulatory proteins much more often than in the neighborhoods of non-regulatory genes, showing that the data provide a sound starting point for studying gene regulation in Synechocystis. Concordantly, we provide several case studies demonstrating that Synergy can be used to find biologically relevant regulatory mechanisms in Synechocystis. Synergy can be used to interactively perform analyses such as gene/motif search, network visualization and motif/function enrichment. Considering the importance of Synechocystis for photosynthesis and biofuel research, we believe that Synergy will become a valuable resource to the research community.
Conflict of interest statement
Figures





Similar articles
-
Integrative analysis of large scale expression profiles reveals core transcriptional response and coordination between multiple cellular processes in a cyanobacterium.BMC Syst Biol. 2010 Aug 2;4:105. doi: 10.1186/1752-0509-4-105. BMC Syst Biol. 2010. PMID: 20678200 Free PMC article.
-
The Transcriptional Landscape of the Photosynthetic Model Cyanobacterium Synechocystis sp. PCC6803.Sci Rep. 2016 Feb 29;6:22168. doi: 10.1038/srep22168. Sci Rep. 2016. PMID: 26923200 Free PMC article.
-
A data integration and visualization resource for the metabolic network of Synechocystis sp. PCC 6803.Plant Physiol. 2014 Mar;164(3):1111-21. doi: 10.1104/pp.113.224394. Epub 2014 Jan 8. Plant Physiol. 2014. PMID: 24402049 Free PMC article.
-
Gene expression patterns of sulfur starvation in Synechocystis sp. PCC 6803.BMC Genomics. 2008 Jul 21;9:344. doi: 10.1186/1471-2164-9-344. BMC Genomics. 2008. PMID: 18644144 Free PMC article.
-
Functional Transcriptomics for Bacterial Gene Detectives.Microbiol Spectr. 2018 Sep;6(5):10.1128/microbiolspec.rwr-0033-2018. doi: 10.1128/microbiolspec.RWR-0033-2018. Microbiol Spectr. 2018. PMID: 30215343 Free PMC article. Review.
Cited by
-
Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803.Nucleic Acids Res. 2017 Nov 16;45(20):11800-11820. doi: 10.1093/nar/gkx860. Nucleic Acids Res. 2017. PMID: 29036481 Free PMC article.
-
Navigating the microalgal maze: a comprehensive review of recent advances and future perspectives in biological networks.Planta. 2024 Oct 5;260(5):114. doi: 10.1007/s00425-024-04543-7. Planta. 2024. PMID: 39367989 Review.
-
Regulation of the scp Genes in the Cyanobacterium Synechocystis sp. PCC 6803--What is New?Molecules. 2015 Aug 12;20(8):14621-37. doi: 10.3390/molecules200814621. Molecules. 2015. PMID: 26274949 Free PMC article.
References
-
- Lee H-S, Vermaas WFJ, Rittmann BE (2010) Biological hydrogen production: prospects and challenges. Trends Biotechnol 28:262–271 Available: http://www.ncbi.nlm.nih.gov/pubmed/20189666 Accessed 2014 Mar 21. - PubMed
-
- Machado IMP, Atsumi S (2012) Cyanobacterial biofuel production. J Biotechnol 162:50–56 Available: http://www.ncbi.nlm.nih.gov/pubmed/22446641 Accessed 2013 May 28. - PubMed
-
- Lindberg P, Park S, Melis A (2010) Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab Eng 12:70–79 Available: http://www.ncbi.nlm.nih.gov/pubmed/19833224 Accessed 2013 Jun 2. - PubMed
-
- Englund E, Pattanaik B, Ubhayasekera SJK, Stensjö K, Bergquist J, et al. (2014) Production of Squalene in Synechocystis sp. PCC 6803. PLoS One 9:e90270 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3953072&tool=p... Accessed 2014 Apr 7. - PMC - PubMed
-
- Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, et al. (2010) CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res 38:D379–81 Available: http://www.ncbi.nlm.nih.gov/pubmed/19880388. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

