Mechanisms of linezolid resistance in staphylococci and enterococci isolated from two teaching hospitals in Shanghai, China
- PMID: 25420718
- PMCID: PMC4245736
- DOI: 10.1186/s12866-014-0292-5
Mechanisms of linezolid resistance in staphylococci and enterococci isolated from two teaching hospitals in Shanghai, China
Abstract
Background: Linezolid is one of the most effective treatments against Gram-positive pathogens. However, linezolid-resistant/intermediate strains have recently emerged in worldwide. The purpose of this study was to analyse the prevalence and resistance mechanisms of linezolid-resistant/intermediate staphylococci and enterococci in Shanghai, China.
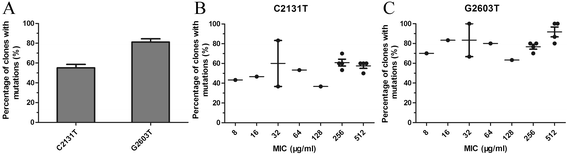
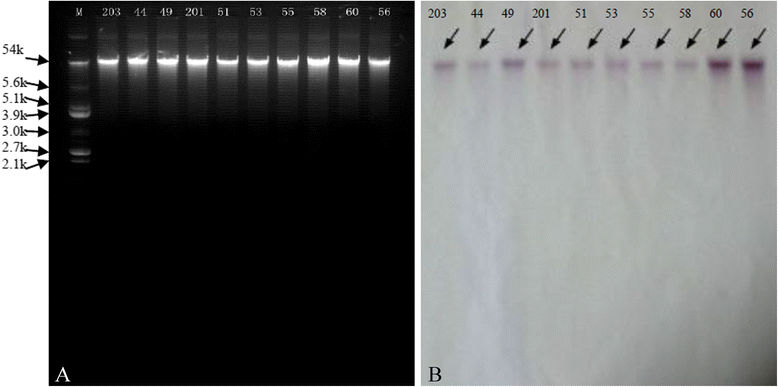
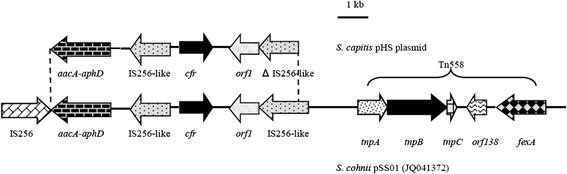
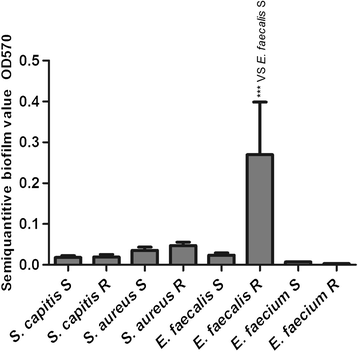
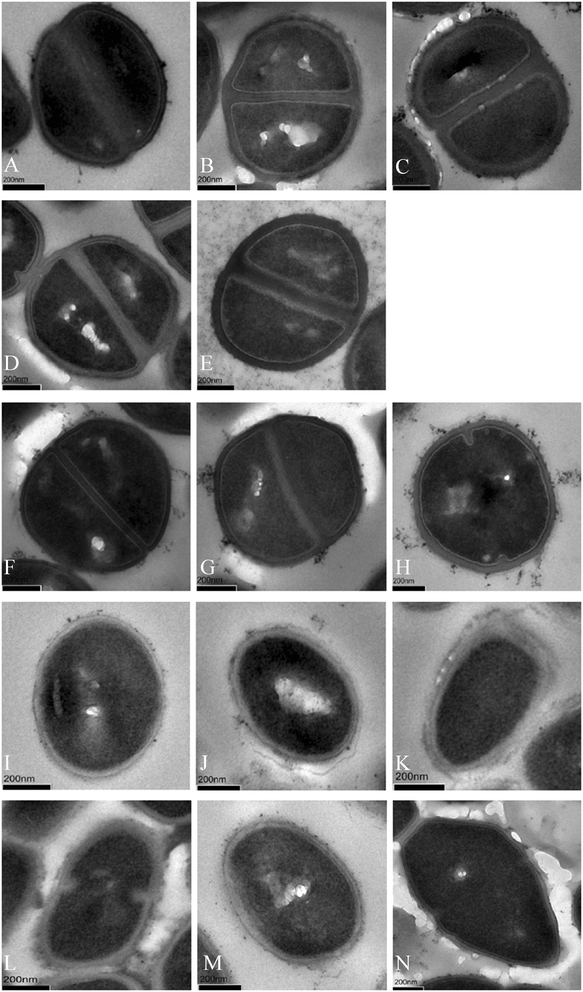
Results: Thirty-two linezolid-resistant/intermediate strains, including 14 Staphylococcus capitis, three Staphylococcus aureus, 14 Enterococcus faecalis and one Enterococcus faecium clinical isolates, were collected in this study which displayed linezolid MICs of 8 to 512 μg/ml, 8-32 μg/ml, 4-8 μg/ml and 4 μg/ml, respectively. All linezolid-resistant S. capitis isolates had a novel C2131T mutation and a G2603T mutation in the 23S rRNA region, and some had a C316T (Arg106Cys) substitution in protein L4 and/or harboured cfr. Linezolid-resistant S. aureus isolates carried a C389G (Ala130Gly) substitution in protein L3, and/or harboured cfr. The cfr gene was flanked by two copies of the IS256-like element, with a downstream orf1 gene. Linezolid-resistant/intermediate enterococci lacked major resistance mechanisms. The semi-quantitative biofilm assay showed that 14 linezolid-resistant E. faecalis isolates produced a larger biofilm than linezolid-susceptible E. faecalis strains. Transmission electron microscopy showed the cell walls of linezolid-resistant/intermediate strains were thicker than linezolid-susceptible strains.
Conclusion: Our data indicated that major resistance mechanisms, such as mutations in 23S rRNA and ribosomal proteins L3 and L4, along with cfr acquisition, played an important role in linezolid resistance. Secondary resistance mechanisms, such as biofilm formation and cell wall thickness, should also be taken into account.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

