Impact of drug resistance-associated amino acid changes in HIV-1 subtype C on susceptibility to newer nonnucleoside reverse transcriptase inhibitors
- PMID: 25421485
- PMCID: PMC4335849
- DOI: 10.1128/AAC.04215-14
Impact of drug resistance-associated amino acid changes in HIV-1 subtype C on susceptibility to newer nonnucleoside reverse transcriptase inhibitors
Abstract
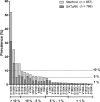
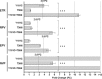
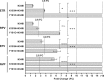
The objective of this study was to assess the phenotypic susceptibility of HIV-1 subtype C isolates, with nonnucleoside reverse transcriptase inhibitor (NNRTI) resistance-associated amino acid changes, to newer NNRTIs. A panel of 52 site-directed mutants and 38 clinically derived HIV-1 subtype C clones was created, and the isolates were assessed for phenotypic susceptibility to etravirine (ETR), rilpivirine (RPV), efavirenz (EFV), and nevirapine (NVP) in an in vitro single-cycle phenotypic assay. The amino acid substitutions E138Q/R, Y181I/V, and M230L conferred high-level resistance to ETR, while K101P and Y181I/V conferred high-level resistance to RPV. Y181C, a major NNRTI resistance-associated amino acid substitution, caused decreased susceptibility to ETR and, to a lesser extent, RPV when combined with other mutations. These included N348I and T369I, amino acid changes in the connection domain that are not generally assessed during resistance testing. However, the prevalence of these genotypes among subtype C sequences was, in most cases, <1%. The more common EFV/NVP resistance-associated substitutions, such as K103N, V106M, and G190A, had no major impact on ETR or RPV susceptibility. The low-level resistance to RPV and ETR conferred by E138K was not significantly enhanced in the presence of M184V/I, unlike for EFV and NVP. Among patient samples, 97% were resistant to EFV and/or NVP, while only 24% and 16% were resistant to ETR and RPV, respectively. Overall, only a few, relatively rare NNRTI resistance-associated amino acid substitutions caused resistance to ETR and/or RPV in an HIV-1 subtype C background, suggesting that these newer NNRTIs would be effective in NVP/EFV-experienced HIV-1 subtype C-infected patients.
Copyright © 2015, American Society for Microbiology. All Rights Reserved.
Figures





References
-
- Geretti AM, Harrison L, Green H, Sabin C, Hill T, Fearnhill E, Pillay D, Dunn D, UK Collaborative Group on HIV Drug Resistance. 2009. Effect of HIV-1 subtype on virologic and immunologic response to starting highly active antiretroviral therapy. Clin Infect Dis 48:1296–1305. doi:10.1086/598502. - DOI - PubMed
-
- Lai MT, Lu M, Felock PJ, Hrin RC, Wang YJ, Yan Y, Munshi S, McGaughey GB, Tynebor RM, Tucker TJ, Williams TM, Grobler JA, Hazuda DJ, McKenna PM, Miller MD. 2010. Distinct mutation pathways of non-subtype B HIV-1 during in vitro resistance selection with nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother 54:4812–4824. doi:10.1128/AAC.00829-10. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

