Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology
- PMID: 25421701
- PMCID: PMC6190593
- DOI: 10.1038/nri3763
Immunity in Drosophila melanogaster--from microbial recognition to whole-organism physiology
Abstract
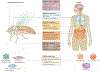
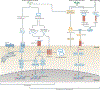
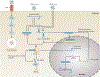
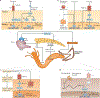
Since the discovery of antimicrobial peptide responses 40 years ago, the fruit fly Drosophila melanogaster has proven to be a powerful model for the study of innate immunity. Early work focused on innate immune mechanisms of microbial recognition and subsequent nuclear factor-κB signal transduction. More recently, D. melanogaster has been used to understand how the immune response is regulated and coordinated at the level of the whole organism. For example, researchers have used this model in studies investigating interactions between the microbiota and the immune system at barrier epithelial surfaces that ensure proper nutritional and immune homeostasis both locally and systemically. In addition, studies in D. melanogaster have been pivotal in uncovering how the immune response is regulated by both endocrine and metabolic signalling systems, and how the immune response modifies these systems as part of a homeostatic circuit. In this Review, we briefly summarize microbial recognition and antiviral immunity in D. melanogaster, and we highlight recent studies that have explored the effects of organism-wide regulation of the immune response and, conversely, the effects of the immune response on organism physiology.
Conflict of interest statement
Competing interests statement
The authors declare no competing interests.
Figures

References
-
- St Johnston D & Nusslein-Volhard C The origin of pattern and polarity in the Drosophila embryo. Cell 68, 201–219 (1992). - PubMed
-
- Padmanabha D & Baker KD Drosophila gains traction as a repurposed tool to investigate metabolism. Trends Endocrinol. Metab 25, 518–527 (2014). - PubMed
-
- Lemaitre B & Hoffmann JA The host defense of Drosophila melanogaster. Annu. Rev. Immunol 25, 697–743 (2007). - PubMed
-
- Vallet-Gely I, Lemaitre B & Boccard F Bacterial strategies to overcome insect defences. Nature Rev. Microbiol 6, 302–313 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

