Comparison of tamoxifen and letrozole response in mammary preneoplasia of ER and aromatase overexpressing mice defines an immune-associated gene signature linked to tamoxifen resistance
- PMID: 25421723
- PMCID: PMC4291054
- DOI: 10.1093/carcin/bgu237
Comparison of tamoxifen and letrozole response in mammary preneoplasia of ER and aromatase overexpressing mice defines an immune-associated gene signature linked to tamoxifen resistance
Abstract
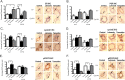
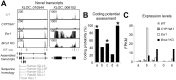
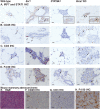
Response to breast cancer chemoprevention can depend upon host genetic makeup and initiating events leading up to preneoplasia. Increased expression of aromatase and estrogen receptor (ER) is found in conjunction with breast cancer. To investigate response or resistance to endocrine therapy, mice with targeted overexpression of Esr1 or CYP19A1 to mammary epithelial cells were employed, representing two direct pathophysiological interventions in estrogen pathway signaling. Both Esr1 and CYP19A1 overexpressing mice responded to letrozole with reduced hyperplastic alveolar nodule prevalence and decreased mammary epithelial cell proliferation. CYP19A1 overexpressing mice were tamoxifen sensitive but Esr1 overexpressing mice were tamoxifen resistant. Increased ER expression occurred with tamoxifen resistance but no consistent changes in progesterone receptor, pSTAT3, pSTAT5, cyclin D1 or cyclin E levels in association with response or resistance were found. RNA-sequencing (RNA-seq) was employed to seek a transcriptome predictive of tamoxifen resistance using these models and a second tamoxifen-resistant model, BRCA1 deficient/Trp53 haploinsufficient mice. Sixty-eight genes associated with immune system processing were upregulated in tamoxifen-resistant Esr1- and Brca1-deficient mice, whereas genes related to aromatic compound metabolic process were upregulated in tamoxifen-sensitive CYP19A1 mice. Interferon regulatory factor 7 was identified as a key transcription factor regulating these 68 immune processing genes. Two loci encoding novel transcripts with high homology to human immunoglobulin lambda-like polypeptide 1 were uniquely upregulated in the tamoxifen-resistant models. Letrozole proved to be a successful alternative to tamoxifen. Further study of transcriptional changes associated with tamoxifen resistance including immune-related genes could expand our mechanistic understanding and lead to biomarkers predictive of escape or response to endocrine therapies.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures



References
-
- Henriksen K.L., et al. (2009). An ER activity profile including ER, PR, Bcl-2 and IGF-IR may have potential as selection criterion for letrozole or tamoxifen treatment of patients with advanced breast cancer. Acta Oncol., 48, 522–531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- NCI U54CA149147/CA/NCI NIH HHS/United States
- CA112176/CA/NCI NIH HHS/United States
- U54 CA149147/CA/NCI NIH HHS/United States
- NIH IG20RR025828/RR/NCRR NIH HHS/United States
- Intramural NIH HHS/United States
- NIH NCI 5P30CA051008/CA/NCI NIH HHS/United States
- BC130883/BC/NCI NIH HHS/United States
- P30 CA051008/CA/NCI NIH HHS/United States
- G20 RR025828/RR/NCRR NIH HHS/United States
- 5T32CA009686-15/CA/NCI NIH HHS/United States
- T32 CA009686/CA/NCI NIH HHS/United States
- U01 CA184902/CA/NCI NIH HHS/United States
- R01 CA112176/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

