HIV-2 interaction with cell coreceptors: amino acids within the V1/V2 region of viral envelope are determinant for CCR8, CCR5 and CXCR4 usage
- PMID: 25421818
- PMCID: PMC4251929
- DOI: 10.1186/s12977-014-0099-3
HIV-2 interaction with cell coreceptors: amino acids within the V1/V2 region of viral envelope are determinant for CCR8, CCR5 and CXCR4 usage
Abstract
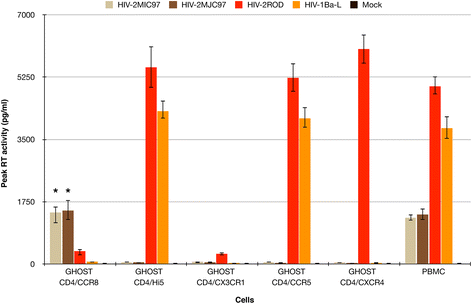
Background: Human immunodeficiency virus 1 and 2 (HIV-1 and HIV-2) use cellular receptors in distinct ways. Besides a more promiscuous usage of coreceptors by HIV-2 and a more frequent detection of CD4-independent HIV-2 isolates, we have previously identified two HIV-2 isolates (HIV-2MIC97 and HIV-2MJC97) that do not use the two major HIV coreceptors: CCR5 and CXCR4. All these features suggest that in HIV-2 the Env glycoprotein subunits may have a different structural organization enabling distinct - although probably less efficient - interactions with cellular receptors.
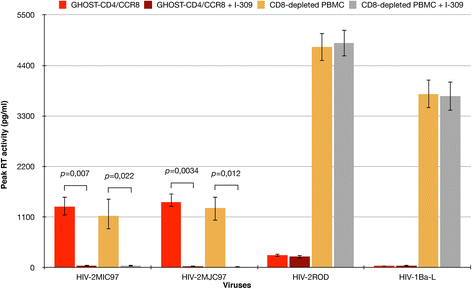
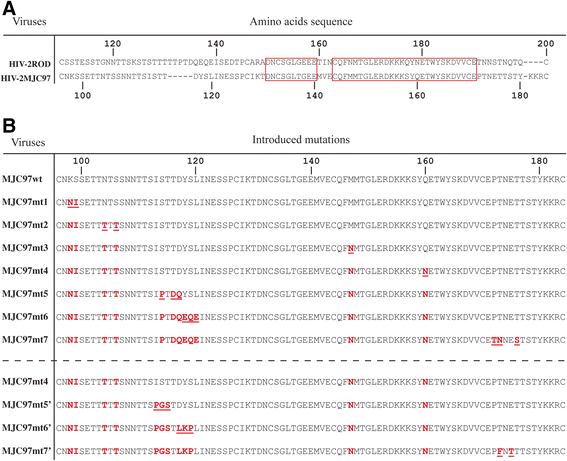
Results: By infectivity assays using GHOST cell line expressing CD4 and CCR8 and blocking experiments using CCR8-specific ligand, I-309, we show that efficient replication of HIV-2MIC97 and HIV-2MJC97 requires the presence of CCR8 at plasma cell membrane. Additionally, we disclosed the determinants of chemokine receptor usage at the molecular level, and deciphered the amino acids involved in the usage of CCR8 (R8 phenotype) and in the switch from CCR8 to CCR5 or to CCR5/CXCR4 usage (R5 or R5X4 phenotype). The data obtained from site-directed mutagenesis clearly indicates that the main genetic determinants of coreceptor tropism are located within the V1/V2 region of Env surface glycoprotein of these two viruses.
Conclusions: We conclude that a viral population able to use CCR8 and unable to infect CCR5 or CXCR4-positive cells, may exist in some HIV-2 infected individuals during an undefined time period, in the course of the asymptomatic stage of infection. This suggests that in vivo alternate molecules might contribute to HIV infection of natural target cells, at least under certain circumstances. Furthermore we provide direct and unequivocal evidence that the usage of CCR8 and the switch from R8 to R5 or R5X4 phenotype is determined by amino acids located in the base and tip of V1 and V2 loops of HIV-2 Env surface glycoprotein.
Figures




Similar articles
-
Differences in molecular evolution between switch (R5 to R5X4/X4-tropic) and non-switch (R5-tropic only) HIV-1 populations during infection.Infect Genet Evol. 2010 Apr;10(3):356-64. doi: 10.1016/j.meegid.2009.05.003. Epub 2009 May 14. Infect Genet Evol. 2010. PMID: 19446658
-
Linkages between HIV-1 specificity for CCR5 or CXCR4 and in vitro usage of alternative coreceptors during progressive HIV-1 subtype C infection.Retrovirology. 2013 Sep 16;10:98. doi: 10.1186/1742-4690-10-98. Retrovirology. 2013. PMID: 24041034 Free PMC article.
-
Coreceptor usage by HIV-1 and HIV-2 primary isolates: the relevance of CCR8 chemokine receptor as an alternative coreceptor.Virology. 2010 Dec 20;408(2):174-82. doi: 10.1016/j.virol.2010.09.020. Epub 2010 Oct 13. Virology. 2010. PMID: 20947116
-
[Chemokine receptors and its importance in the replication cycle of human immunodeficiency virus: clinical and therapeutic implications].Acta Med Port. 2008 Sep-Oct;21(5):497-504. Epub 2009 Jan 16. Acta Med Port. 2008. PMID: 19187693 Review. Portuguese.
-
Molecular mechanisms of HIV entry.Adv Exp Med Biol. 2012;726:223-42. doi: 10.1007/978-1-4614-0980-9_10. Adv Exp Med Biol. 2012. PMID: 22297516 Review.
Cited by
-
CRISPR Technology in Gene-Editing-Based Detection and Treatment of SARS-CoV-2.Front Mol Biosci. 2022 Jan 11;8:772788. doi: 10.3389/fmolb.2021.772788. eCollection 2021. Front Mol Biosci. 2022. PMID: 35087864 Free PMC article. Review.
-
HIV capsids: orchestrators of innate immune evasion, pathogenesis and pandemicity.J Gen Virol. 2025 Jan;106(1):002057. doi: 10.1099/jgv.0.002057. J Gen Virol. 2025. PMID: 39804283 Free PMC article. Review.
-
Cell-to-Cell Transmission of HIV-1 and HIV-2 from Infected Macrophages and Dendritic Cells to CD4+ T Lymphocytes.Viruses. 2023 Apr 22;15(5):1030. doi: 10.3390/v15051030. Viruses. 2023. PMID: 37243118 Free PMC article.
-
HIV-2 Neutralization Sensitivity in Relation to Co-Receptor Entry Pathways and Env Motifs.Int J Mol Sci. 2022 Apr 26;23(9):4766. doi: 10.3390/ijms23094766. Int J Mol Sci. 2022. PMID: 35563157 Free PMC article.
-
A genotypic method for determining HIV-2 coreceptor usage enables epidemiological studies and clinical decision support.Retrovirology. 2016 Dec 20;13(1):85. doi: 10.1186/s12977-016-0320-7. Retrovirology. 2016. PMID: 27998283 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

