Hepatitis B virus HBx protein interactions with the ubiquitin proteasome system
- PMID: 25421893
- PMCID: PMC4246244
- DOI: 10.3390/v6114683
Hepatitis B virus HBx protein interactions with the ubiquitin proteasome system
Abstract
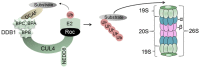
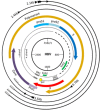
The hepatitis B virus (HBV) causes acute and chronic hepatitis, and the latter is a major risk factor for the development of hepatocellular carcinoma (HCC). HBV encodes a 17-kDa regulatory protein, HBx, which is required for virus replication. Although the precise contribution(s) of HBx to virus replication is unknown, many viruses target cellular pathways to create an environment favorable for virus replication. The ubiquitin proteasome system (UPS) is a major conserved cellular pathway that controls several critical processes in the cell by regulating the levels of proteins involved in cell cycle, DNA repair, innate immunity, and other processes. We summarize here the interactions of HBx with components of the UPS, including the CUL4 adaptor DDB1, the cullin regulatory complex CSN, and the 26S proteasome. Understanding how these protein interactions benefit virus replication remains a challenge due to limited models in which to study HBV replication. However, studies from other viral systems that similarly target the UPS provide insight into possible strategies used by HBV.
Figures


References
-
- Voutsadakis I.A. Ubiquitin, ubiquitination and the ubiquitin-proteasome system in cancer. Atlas Genet. Cytogenet. Oncol. Haematol. 2010;14:1088–1099.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

