Functions of the proteasome on chromatin
- PMID: 25422899
- PMCID: PMC4279168
- DOI: 10.3390/biom4041026
Functions of the proteasome on chromatin
Abstract
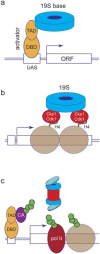
The proteasome is a large self-compartmentalized protease complex that recognizes, unfolds, and destroys ubiquitylated substrates. Proteasome activities are required for a host of cellular functions, and it has become clear in recent years that one set of critical actions of the proteasome occur on chromatin. In this review, we discuss some of the ways in which proteasomes directly regulate the structure and function of chromatin and chromatin regulatory proteins, and how this influences gene transcription. We discuss lingering controversies in the field, the relative importance of proteolytic versus non-proteolytic proteasome activities in this process, and highlight areas that require further investigation. Our intention is to show that proteasomes are involved in major steps controlling the expression of the genetic information, that proteasomes use both proteolytic mechanisms and ATP-dependent protein remodeling to accomplish this task, and that much is yet to be learned about the full spectrum of ways that proteasomes influence the genome.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

