Predictive gene signatures: molecular markers distinguishing colon adenomatous polyp and carcinoma
- PMID: 25423035
- PMCID: PMC4244109
- DOI: 10.1371/journal.pone.0113071
Predictive gene signatures: molecular markers distinguishing colon adenomatous polyp and carcinoma
Abstract
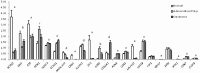
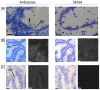
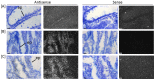
Cancers exhibit abnormal molecular signatures associated with disease initiation and progression. Molecular signatures could improve cancer screening, detection, drug development and selection of appropriate drug therapies for individual patients. Typically only very small amounts of tissue are available from patients for analysis and biopsy samples exhibit broad heterogeneity that cannot be captured using a single marker. This report details application of an in-house custom designed GenomeLab System multiplex gene expression assay, the hCellMarkerPlex, to assess predictive gene signatures of normal, adenomatous polyp and carcinoma colon tissue using archived tissue bank material. The hCellMarkerPlex incorporates twenty-one gene markers: epithelial (EZR, KRT18, NOX1, SLC9A2), proliferation (PCNA, CCND1, MS4A12), differentiation (B4GANLT2, CDX1, CDX2), apoptotic (CASP3, NOX1, NTN1), fibroblast (FSP1, COL1A1), structural (ACTG2, CNN1, DES), gene transcription (HDAC1), stem cell (LGR5), endothelial (VWF) and mucin production (MUC2). Gene signatures distinguished normal, adenomatous polyp and carcinoma. Individual gene targets significantly contributing to molecular tissue types, classifier genes, were further characterised using real-time PCR, in-situ hybridisation and immunohistochemistry revealing aberrant epithelial expression of MS4A12, LGR5 CDX2, NOX1 and SLC9A2 prior to development of carcinoma. Identified gene signatures identify aberrant epithelial expression of genes prior to cancer development using in-house custom designed gene expression multiplex assays. This approach may be used to assist in objective classification of disease initiation, staging, progression and therapeutic responses using biopsy material.
Conflict of interest statement
Figures




References
-
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, et al. (2008) v1.2, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 10 [Internet] Lyon, France: International Agency for Research on Cancer, (2010) Available from: http://globocan.iarc.fr.
-
- Steele RJ, McClements PL, Libby G, Black R, Morton C, et al. (2009) Results from the first three rounds of the Scottish demonstration pilot of FOBT screening for colorectal cancer. Gut 58:530–553. - PubMed
-
- Hanahan D, Weinberg RA (2000) The Hallmarks of cancer. Cell 100, 57–70. - PubMed
-
- Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144:646–674. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

